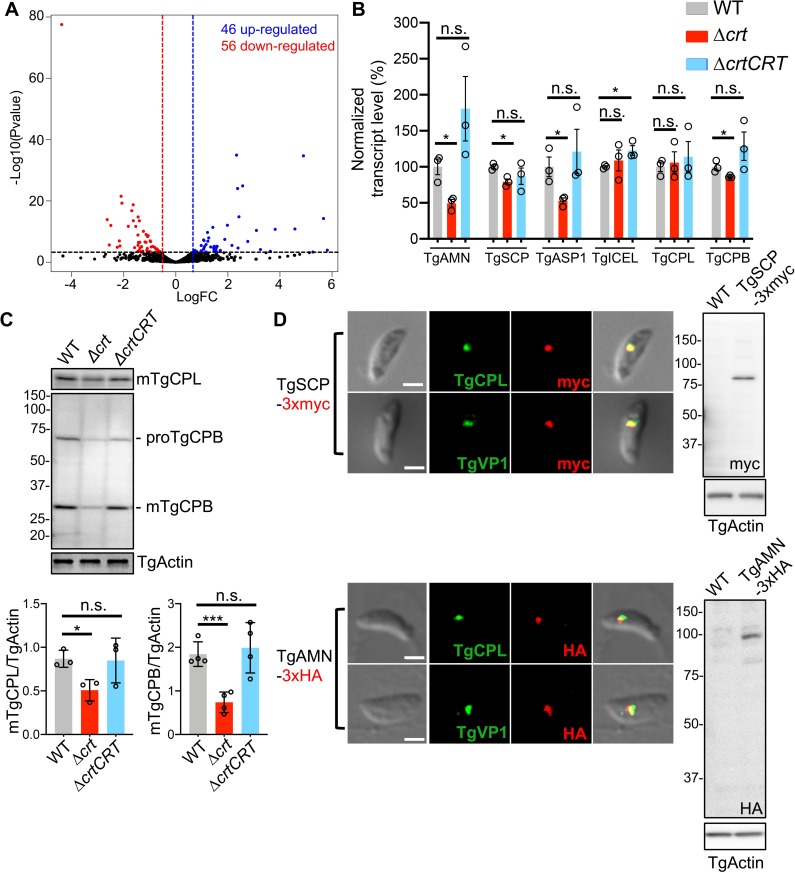
Fig 5. The transcript and protein abundances of several VAC-residing proteases were decreased in Δcrt.
(A) RNA-Seq was performed in WT and Δcrt parasites. Each sample was sequenced in duplicate for statistical comparison. A volcano plot was used to summarize the genes with altered transcription greater than 1.5-fold and with statistical significance less than 0.05 in the Δcrt mutant relative to the WT strain. Forty-six and fifty-six genes labeled in the blue and red dots became up- and down-regulated in the Δcrt mutant, respectively. The blue and red dash lines represent the borderline of 1.5-fold change in gene transcripts, and the genes above the black dash line had p values of statistical significance below 0.05. (B) qPCR was used to validate 4 down-regulated proteases that were identified by RNA-Seq analysis, along with two known VAC proteases, TgCPL and TgCPB. TgAMN, TgSCP, TgASP1, and TgCPB displayed down-regulated transcription in Δcrt parasites compared to WT. The qPCR assay was repeated in three technical replicates per biological replicate in a total of three biological replicates. Data shown in the figure are represented as mean ± SEM. The fold change of individual genes was calculated using a ΔΔCT (cycle threshold) method for relative quantification by normalization against WT values using TgActin as a normalization control. The variation shown in WT parasites was determined by normalization of individual CT values per replicate against the input of total RNA. (C) The steady protein abundances of TgCPL and TgCPB were quantified in the lysates of parasites by immunoblotting. The protein levels of TgCPL and TgCPB in the Δcrt mutant were reduced by ~40% and 60%, respectively, compared to WT parasites. Immunoblotting assays were repeated in 3–4 replicates for statistical comparison. (D) TgSCP and TgAMN were endogenously tagged with 3xmyc and 3xHA, respectively, at their C-termini. The expression of the epitope-tagged proteins was confirmed by immunoblotting. The parasites were co-stained with antibodies recognizing their respective epitope tags as well as the VAC and ELC markers. Both TgSCP and TgAMN were localized in the VAC and ELC by immunofluorescence. Statistical significance was performed using unpaired two-tailed Student’s t-test. *, p<0.05; ***, p<0.001; n.s., not significant.