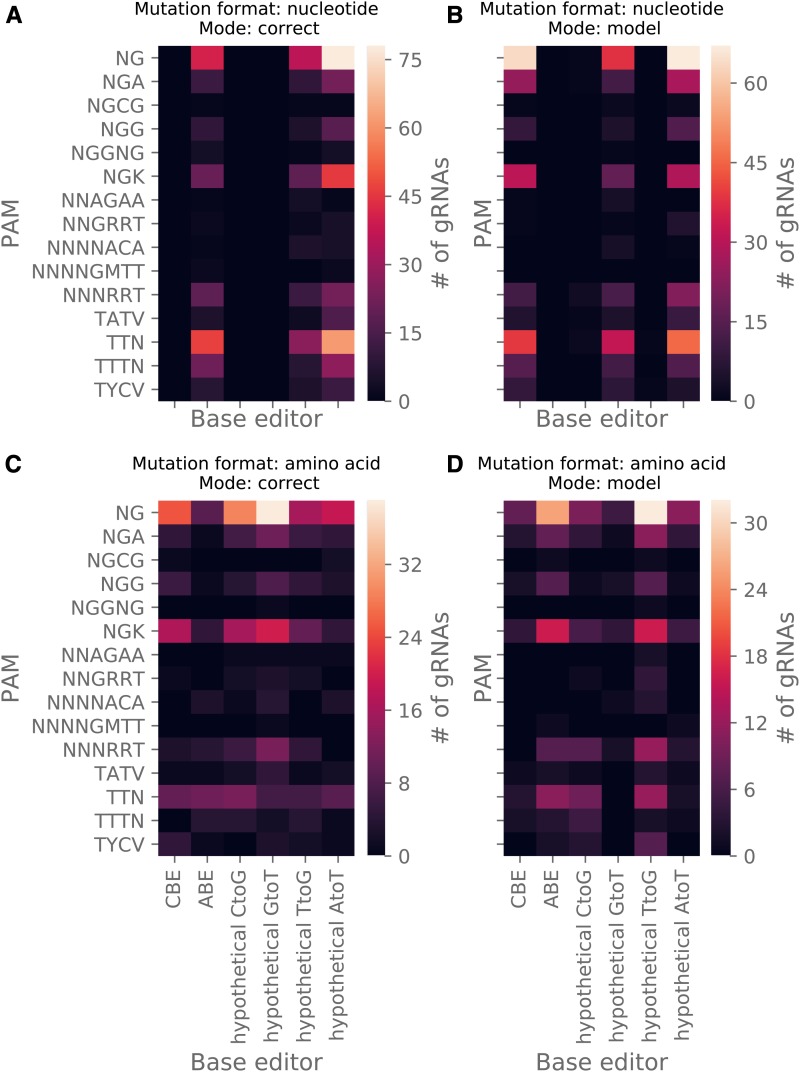
Figure 2.
Demonstrative analysis of gRNAs designed with custom base editors and PAM recognition sequences. To demonstrate the utility of beditor in utilizing custom base editors and PAM recognition sequences, sets of 1000 randomly assigned mutations in S. cerevisiae (see Supplemental methods) were analyzed in two mutation formats (nucleotide or amino acid) and two modes of mutagenesis (“model” and “correct”) (each combination is shown in parts A–D). In each heatmap, number of gRNAs designed by each combination of a base editor (in columns) and PAM recognition sequence (in rows) is shown.