Abstract.
Surface waters are an unappreciated reservoir of antimicrobial resistance (AMR). Poor sanitation brings different species of environmental bacteria into contact, facilitating horizontal gene transfer. To investigate the role of surface waters as potential reservoirs of AMR, we studied the point prevalence of fecal contamination, AMR genes, and Enterobacteriaceae in an urban lake and rural river system in Northeast Brazil in comparison with a lake and sewer system in Northeast Ohio in the United States. Surface water samples were examined for evidence of human fecal contamination using microbial source tracking and screened for plasmid-mediated fluoroquinolone resistance and carbapenemase genes. Enterobacteriaceae were detected using selective agar followed by antimicrobial susceptibility testing and detection of AMR genes by microarray, and classified by repetitive sequence–based polymerase chain reaction and multilocus sequence typing. Concentrations of human fecal bacteria in the Brazilian urban lake and sewage in Northeast Ohio were similarly high. Filtered water samples from the Brazilian urban lake, however, showed the presence of blaOXA-48, blaKPC, blaVIM-2, qnrS, and aac(6′)-lb-cr, whereas only blaVIM-2 was identified in raw sewage from Northeast Ohio. From the Brazilian urban lake, 85% of the Enterobacteriaceae (n = 40) cultured were resistant to at least one clinically important antibiotic, including ST131 Escherichia coli harboring the extended-spectrum beta-lactamase CTX-M. Although two isolates demonstrated polymyxin resistance, mcr-1/2 was not detected. Our findings indicate that surface waters in an urban Brazilian site can serve as an environmental reservoir of AMR and that improving wastewater treatment and sanitation generally may ameliorate AMR dissemination.
INTRODUCTION
The emergence of antimicrobial resistance (AMR) in the environment is the result of a complex interaction between biology, human behavior, and geography that is often impacted by selective pressure from the use of antibiotics.1 Surface water, conventionally defined as water on the surface of the planet, such as in a river, lake, wetland, or ocean, is likely an important reservoir for the maintenance and transmission of antibiotic-resistant coliform and non-coliform bacteria.2 Similarly, the presence of AMR genes and microbial contaminants has been documented in groundwater.3 Although AMR determinants can occur naturally and various species of bacteria have intrinsic resistance to different antibiotics, contamination of the environment with human and animal feces likely contributes to the dissemination of AMR.4–6
The prevalence of AMR in the natural environment may not precisely correlate with antibiotic use in humans, but rather is influenced by other social, economic, and sanitation factors.1 The prevalence of infections and the use of antibiotics follows the distribution of people and animals so that migration7–10 and dense urban environments provide conditions that may promote AMR transmission.11 In low- and middle-income countries, rapid urbanization has led to crowding and poor sanitation in affecting aquatic environments that bring human, animal, and environmental bacteria into direct contact, facilitating horizontal gene transfer of AMR genes.12,13
The WHO has identified Enterobacteriaceae resistant to carbapenems, extended-spectrum cephalosporins, or fluoroquinolones as priority pathogens.14,15 These multidrug-resistant and extensively drug-resistant organisms are major contributors to human morbidity and mortality and can be distributed widely, from person-to-person, between animals and humans, and from both to the environment. Furthermore, few therapeutic options are available to treat these resistant pathogens. Even antibiotics reserved for the treatment of extensively resistant Gram-negative bacteria are affected: resistance to polymyxins has been linked to genes contained in a plasmid from commensal Escherichia coli in food animals, which has disseminated around the world in both animals and humans.16
Our aim was to conduct a point prevalence study to investigate whether surface water sampled from a typical urban setting in Northeast Brazil is a potential source of AMR. We used as comparison surface waters from geographic and socioeconomic communities which differ in land use, climate, antibiotic consumption, and management of wastewater. Specific sites included a rural river system in Brazil adjacent to cattle pastures, as well as an urban lake and a metropolitan wastewater treatment plant in the United States. Our hypothesis was that differences in fecal contamination determined the presence of AMR genes and bacteria of critical importance to human health. We quantified the degree of contamination with human and ruminant feces; screened surface water samples for AMR genes; detected the presence of Enterobacteriaceae resistant to carbapenems, extended-spectrum cephalosporins, fluoroquinolones, and polymyxins; and identified their genetic determinants of resistance.
MATERIALS AND METHODS
Study sites.
The Jiquiriçá and Brejões rivers intersect within the rural village of Jenipapo, located in the Brazilian state of Bahia. The community consists of 175 households and 617 residents whose primary occupation is small-scale agriculture. The village is 263 km southeast of Salvador, the state capital. Nearly all households receive piped water treated at a local plant and have a flush toilet, but only 43% of households have a septic tank.17 The remaining residents discharge their wastewater directly into the rivers.
The Brazilian urban lake, Dique do Cabrito, is a small artificial lake (surface area of approximately 74,000 m2) located downstream of the Cobre River and adjacent to Bahia de Todos os Santos in the Atlantic Ocean. Dique do Cabrito is surrounded by densely populated neighborhoods in Salvador, a major metropolitan area and Brazil’s 4th most populous city with 2.9 million residents. It is estimated that 87% of households have adequate sewage disposal, divided between the municipal system and septic tanks. The remainder of residents discharge their wastewater into several bodies of fresh water, including the Dique do Cabrito.18 Dique do Cabrito, however, does not receive effluent from any hospital.
The urban lake in the United States is Lower Shaker Lake (surface area of approximately 71,200 m2), an artificial lake that is part of the Doan Brook watershed system. It is located in the city of Shaker Heights, an inner suburb of Cleveland, OH, with a population of 27,000. This site was selected because it shares common elements with the Brazilian urban site, such as its size and being within a residential community. This body of water does not directly receive wastewater but had a combined storm water overflow and sanitary sewer system. Efforts to upgrade this system are underway.
A pretreatment sewage sample was obtained from a water treatment facility in Northeast Ohio. The facility serves Cleveland and the surrounding suburbs and processes an average of 120 million gallons of water per day (www.neorsd.org/about/southerly/). Cleveland’s metropolitan area contains a population of approximately 2 million.
Water collection.
We obtained water samples from separate locations within each site. A total of 10 locations along the course of the rural river system in Brazil and two locations from the community’s piped water system were surveyed in July 2015. The piped water from the rural community is drawn from a surface collection site and is chlorinated and filtered before pumping into storage tanks. Samples from the urban lake in Brazil were obtained from 15 locations around its perimeter, also in July 2015. Samples from five locations in the perimeter of the urban lake and untreated sewer wastewater in Northeast Ohio were collected in October 2015. All collections were made between 8 am and 12 pm local time in clean 500-mL bottles for filtration and sterile 50-mL conical tubes for culture. The samples were placed in a cooler with ice packs until processing. Water was processed for coliform quantification and filtration within 2 hours after collection. Samples for culture and sensitivity assays were stored at 4°C for an additional 24 hours before processing.
Coliform counts.
Aliquots (1 mL) were removed from water collected at each site and mixed with Coliscan Easygel media (Micrology Laboratories, Goshen, IN). After incubation for 24–48 hours at 30°C, colonies were counted manually up to 1,000 CFU/mL. Escherichia coli was differentiated from other thermotolerant coliforms by their blue/purple or pink color, respectively.19
Detection of antibiotic resistance genes in water samples.
We screened for the presence of common plasmid-mediated quinolone resistance (PMQR) (qnrA, qnrS, and aac(6′)-lb-cr) and carbapenemase (blaNDM, blaSPM, blaIMP, blaOXA-48, blaKPC, and blaVIM) genes in DNA from water samples using PCR. We also tested Enterobacteriaceae isolates for the presence of plasmid-mediated colistin resistance genes mcr-1 and mcr-2. Primers used for amplification are summarized in Table 1.
Table 1.
Primers used in this study
| Target | Sequence (Forward, 5′-3′) | Ref. |
|---|---|---|
| Bac32F (Bacteroides spp. and Prevotella spp.) | AACGCTAGCTACAGGCTT | 52 |
| HF183F (Human (HF8) cluster Bacteroides spp.) | ATCATGAGTTCACATGTCCG | 53 |
| Lachno2F (Lachnospiraceae spp. Human cluster) | TTCGCAAGAATGAAACTCAAAG | 21 |
| BacR_f (ruminant Bacteroidetes spp.) | GCGTATCCAACCTTCCCG | 21 |
| blaKPC (Klebsiella pneumoniae carbapenemase gene) | TGTCACTGTATCGCCGTC | 53 |
| blaIMP (metallo-beta-lactamase gene) | GAAGGCGTTTATGTTCATAC | 54 |
| blaVIM (Verona integron–mediated metallo-beta-lactamse gene) | GTTTGGTCGCATATCGCAAC | 1 |
| blaNDM (New Delhi metallo-beta-lactamase gene) | GCAGCTTGTCGGCCATGCGGGC | 54 |
| blaOXA-48-like (carbapenemase gene) | GCGTGGTTAAGGATGAACAC | 54 |
| blaSPM (Sao Paulo metallo-beta-lactamase gene) | AAAATCTGGGTACGCAAACG | 54 |
| qnrS (fluoroquinolone plasmid-mediated resistance gene) | GGAAACCTACAATCATACATA | 55 |
| qnrA (fluoroquinolone plasmid-mediated resistance gene) | GATAAAGTTTTTCAGCAAGAGG | 56 |
| aac(6′)-lb-cr (fluoroquinolone plasmid-mediated resistance gene) | TGACCTTGCGATGCTCTATG | 57 |
| mcr-1 (colistin/polymyxin B resistance gene) | ATGATGGCAGCATACTTCTGTGTGG | 22 |
| mcr-2 (colistin/polymyxin B resistance gene) | TGTTGCTTGTGCCGATTGGA | 19 |
Microbial source tracking.
From each site, 500 mL of water was filtered through a 47-mm-diameter mixed cellulose filter with 0.22-µm sized pores (EMD Millipore Corporation, Billerica, MA) using a Nalgene filter funnel unit (Thermo Scientific, Waltham, MA). The filters were thoroughly dried and stored at −20°C until processing. Frozen filters were broken into small fragments with a sterile metal spatula, DNA was extracted with phenol–chloroform, and then followed by treatment with hexadecyltrimethylammonium bromide to remove PCR inhibitors.
Non-fluorophore–labeled primer sets (Table 1) based on the 16S rRNA gene were used to detect general Bacteroidales spp., as well as human Bacteroidales and ruminant Bacteroidales, which are highly specific for tracking the source of human and ruminant fecal pollution.20 To measure fecal contamination based on these markers, quantitative PCR was performed using FastStart Universal SYBR Green Master Mix (Roche Diagnostics Corporation, Indianapolis, IN). Standard curves were generated during each run and consisted of an amplicon produced from either a plasmid containing the target sequence or ruminant fecal samples. Each assay was performed in duplicate and the results were averaged.21
Detection of antibiotic-resistant Enterobacteriaceae.
To screen for Enterobacteriaceae resistant to fluoroquinolones, extended-spectrum cephalosporins, and carbapenems, we centrifuged 10 mL of water at 5,000 × g for 5 minutes, resuspended the pellet in tryptic soy broth (Becton-Dixon, Franklin Lakes, NJ), incubated for 24 hours at 36°C, followed by a culture on a MacConkey agar plate (Oxoid, Basingstoke Hampshire, United Kingdom) with discs for ciprofloxacin (5 µg/mL), cefotaxime (30 µg/mL), and meropenem (10 µg/mL) (Laborclin, Pinhais, Brazil). After incubation for 18–24 hours at 36°C, distinct colonies within the inhibition zone were isolated and further identified.
Once initial biochemical analyses using Enterokit B (Probac do Brasil, São Paulo, Brazil) were performed, confirmation of bacterial species, antibiotic susceptibility, and extended-spectrum β-lactamases (ESBL) phenotypical screening were performed with VITEK-2 (bioMérieux, Marcy l’Etoile, France) and MicroScan (Beckman Coulter, Inc., Brea, CA). Results were interpreted following the 2015 Clinical and Laboratory Standards Institute (CLSI) guidelines. Minimum inhibitory concentrations of polymyxin B were determined using the broth dilution method and interpreted following the CLSI guidelines.22
To determine the types of beta-lactamase genes present in ESBL-positive Enterobacteriaceae, we used a microarray assay based on match ligation-mediated amplification, hybridization, and detection.23 The version we used (CT101XL; Check-Points B.V., Wageningen, the Netherlands) detected genes for the following beta-lactamase, among others: KPC, NDM, and CTX-M-1 group (which includes CTX-M-15); CTX-M-2 group; CTX-M-8 and -25 group; and CTX-M-9 group (which includes CTX-M-14). We also screened with PCR all Enterobacteriaceae isolated in this study for the presence of the following carbapenemase genes: blaOXA-48-like, blaVIM, and blaIMP.22
Genetic typing with repetitive sequence–based PCR and multilocus sequence typing.
Relatedness of E. coli and Klebsiella pneumoniae isolates was determined with repetitive sequence–based PCR (rep-PCR) using a commercial automated system (DiversiLab; bioMérieux). Isolates with rep-PCR similarity ≥ 95% were classified as clonally related.24 We used the band pattern generated by DiversiLab to identify E. coli ST131, as previously described.25
Multilocus sequence typing (MLST) was carried out to determine the sequence types of E. coli and K. pneumoniae isolates that were clonally related according to rep-PCR. Escherichia coli isolates were typed using the sequences of housekeeping genes dinB, icdA, pabB, polB, putP, trpA, trpB, and uidA and K. pneumoniae based on the sequencing of gapA, infB, mdh, pgi, phoE, rpoB, and tonB, following the schemes developed at the Institute Pasteur (http://bigsdp.pasteur.fr/).
RESULTS
Coliform counts.
Samples from the Brazilian urban lake averaged 1.3 × 105 coliforms/site (range 1.9 × 104 to > 4 × 105) and 6.9 × 104 E. coli/site (range 4 × 102 to 2 × 105). By contrast, samples from the Brazilian rural river system ranged between 3 × 102 to 4 × 103 coliforms/mL and 4 × 101 to 1 × 103 E. coli/mL. Small numbers of coliforms but no E. coli were isolated from water samples obtained directly from the treatment plant. For the urban lake in the United States, coliforms were between 1 × 101 to 7 × 103 CFU/mL and none were E. coli. Wastewater samples from the sewer system contained > 4 × 105 CFU/mL for total coliforms and > 2 × 105 CFU/mL for E. coli.
Microbial source tracking.
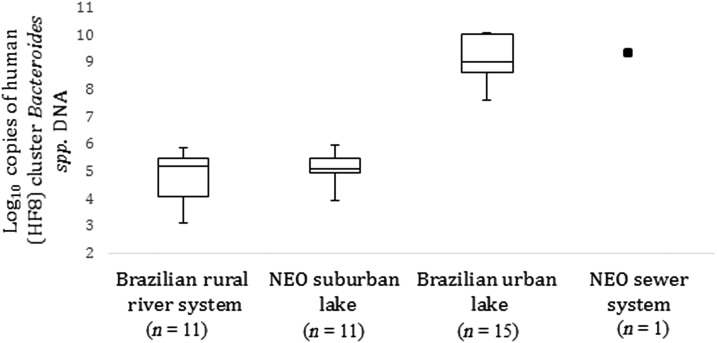
In the Brazilian urban lake, the concentration of human Bacteroidales averaged 4.7 × 109 copies of DNA/100 mL in the 15 collection sites (range 4.8 × 107 to 1.4 × 1010). The highest concentration of human Bacteroidales occurred in the northern half of the lake, as represented in Figure 1, and was comparable with concentrations found in the sample of raw sewage from Northeast Ohio (1.3 × 1010 copies of DNA/100 mL). By contrast, the concentration of fecal bacteria of human origin in samples from the Brazilian rural river system averaged 2.1 × 105 copies of DNA/100 mL of water (range 0–7.6 × 105), whereas the urban lake in Northeast Ohio averaged 5.2 × 104 copies of DNA/100 mL (range 8.7 × 103 to 1.7 × 105) (Figure 2). Only the samples from the Brazilian rural river system contained ruminant Bacteroidales, with a range between 0 and 1.2 × 104 copies of DNA/100 mL.
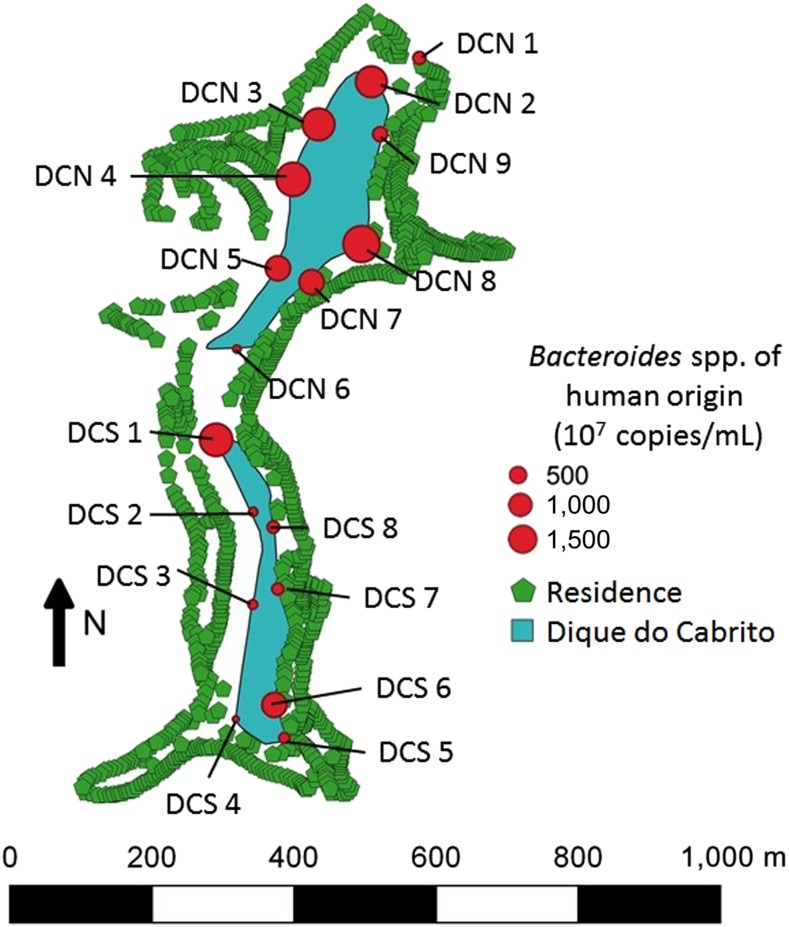
Figure 1.
Map of Dique do Cabrito identifying the number of copies of human Bacteroides spp. at each survey site. DCN indicates a site in the northern half of Dique do Cabrito and DCS indicates a southern site. This figure appears in color at www.ajtmh.org.
Figure 2.
Comparison between study sites of human Bacteroides quantities determined by microbial source tracking. Presents copies of DNA of human (HF8) cluster Bacteroides spp. identified by quantitative PCR in samples from filtered waters. Only one sample of raw sewage was obtained from a sewer system in Northeast Ohio (NEO). A sample from piped water from the Brazilian rural river system did not contain human (HF8) cluster Bacteroides spp. and is not represented.
Detection of antibiotic resistant Enterobacteriaceae.
In all, we identified 40 Enterobacteriaceae isolates resistant to ciprofloxacin, cefotaxime, or meropenem. Using VITEK, the isolates were further identified as E. coli (28), K. pneumoniae (10), and Enterobacter cloacae (two). Most (85%) were from the Brazilian urban lake (23 E. coli, nine K. pneumoniae, and two E. cloacae). Despite similar concentrations of human feces, few were found in raw sewage from the United States (four E. coli and one K. pneumoniae) and even fewer from the Brazilian rural river system (one E. coli). None were cultured from the urban lake in the United States.
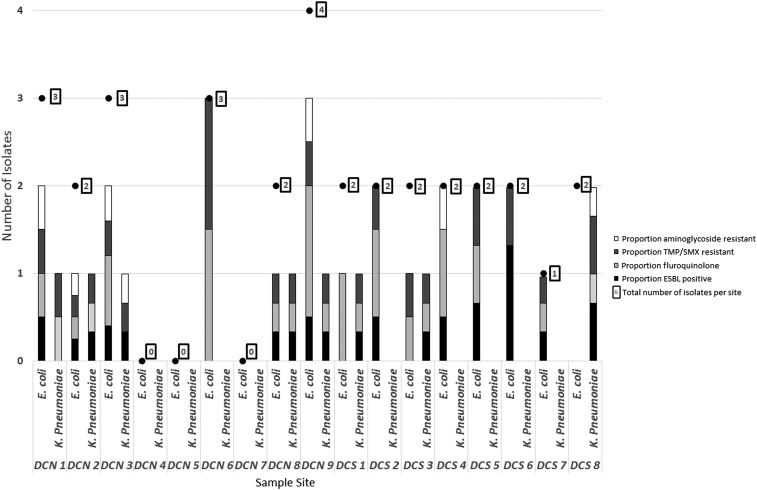
Enterobacteriaceae resistant to fluoroquinolones, aminoglycosides, trimethoprim/sulfamethoxazole (TMP/SMX), or extended-spectrum cephalosporins were identified more frequently from the urban lake in Brazil than any other location (Table 2, Figure 3). Of the isolates cultured from the Brazilian lake, 82.4% were confirmed to be resistant to fluoroquinolones, 73.5% to TMP/SMX, and 29.4% to aminoglycosides. Production of ESBL occurred in 62.5%. Furthermore, multidrug resistance occurred in 76.5% of isolates.26 Resistance to polymyxin B was detected in one isolate of E. coli and E. cloacae each.
Table 2.
Enterobacteriaceae isolates from the Brazilian urban lake
| Escherichia coli, N = 23 | Klebsiella pneumoniae, N = 9 | Enterobacter cloacae, N = 2 | |
|---|---|---|---|
| Extended-spectrum β-lactamases positive* | 12 (52%) | 8 (89%) | N/A |
| Carbapenem resistant* | 0 | 0 | 0 |
| Fluoroquinolone resistant* | 21 (91%) | 7 (78%) | 0 |
| Trimethoprim/sulfamethoxazole resistant* | 14 (61%) | 9 (100%) | 2 (100%) |
| Aminoglycoside resistant* | 6 (26%) | 2 (22%) | 1 (50%) |
| Polymyxin B resistant† | 1 (4%) | 0 | 1 (50%) |
| MDR∥ | 18 (78%) | 8 (89%) | 0 |
| blaKPC‡,§ | 0 | 0 | 0 |
| blaCTX-M‡,§ | 12 (52%) | 5 (55%) | 0 |
| blaTEM‡ | 4 (17%) | 1 (11%) | 0 |
| blaSHV‡ | 0 | 5 (55%) | 0 |
| qnrS§ | 1 (7%) | 0 | 0 |
| qnrA§ | 0 | 0 | 0 |
| aac(6′)-lb-cr§ | 0 | 2 (22%) | 0 |
| mcr-1 or mcr-2§ | 0 | 0 | 0 |
* Results obtained by Vitek-2.
† Results obtained by broth macrodilution.
‡ Results obtained by Check-Points.
§ Results obtained by PCR.
∥ Multidrug resistance (MDR) indicates isolates non-susceptible to ≥ 1 agent in > 3 antimicrobial categories, according to definitions by Magiorakos et al.26
Figure 3.
Resistance phenotypes of Escherichia coli and Klebsiella pneumoniae isolated from Dique do Cabrito. The relative contributions of each of the four resistance phenotypes listed are represented by a single bar per species and location. DCN indicates a site in the northern half of Dique do Cabrito and DCS indicates a southern site. ESBL = extended-spectrum beta-lactamase; TMP/SMX = trimethoprim/sulfamethoxazole.
Of the E. coli from the raw sewage in the United States, one isolate demonstrated ESBL production and was resistant to fluoroquinolones, aminoglycosides, and TMP/SMX. Another isolate was resistant to cefepime and TMP/SMX, and two were resistant to only TMP/SMX. The K. pneumoniae isolate from raw sewage was only resistant to ampicillin, and the isolate of E. coli from the Brazilian rural river system was only resistant to fluoroquinolones.
Among 20 ESBL-producing Enterobacteriaceae isolated from the urban lake in Brazil, blaCTX-M was the gene most frequently detected (80%), occurring in 92% (11/12) of E. coli and 63% (5/8) of K. pneumoniae. We found that CTX-M-9 and CTX-M-1 were similarly present in E. coli and K. pneumoniae. By contrast, blaTEM was detected in 33% (4/12) of E. coli and 13% (1/8) of K. pneumoniae, whereas blaSHV occurred in 25% (2/8) of K. pneumoniae only. The E. coli isolate from the raw sewage sample contained blaCTX-M. We did not detect blaKPC, blaOXA-48, blaNDM, blaIMP, nor blaVIM in any of the Enterobacteriaceae isolates from this survey. Only fluoroquinolone-resistant Enterobacteriaceae isolated from samples from the urban lake in Brazil contained PMQR genes, including qnrS in one isolate of E. coli and aac(6′)-lb-cr in two isolates of K. pneumoniae. The mcr-1 or mcr-2 genes, coding for resistance to polymyxins, were not detected in any of the isolates, including the two colistin-resistant isolates.
Genetic typing with repetitive sequence–based PCR and multilocus sequence typing.
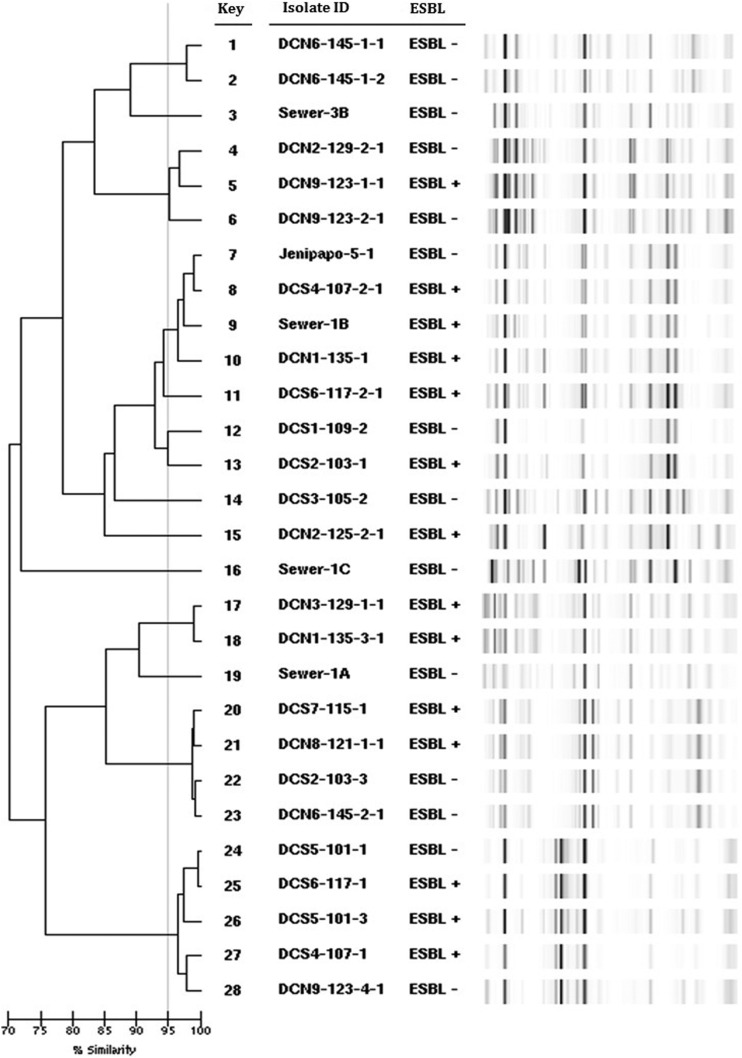
Analysis of genetic relatedness using rep-PCR revealed that 22 of the 28 E. coli grouped into seven clusters (containing two to five isolates each) (Figure 4). According to their rep-PCR pattern, five isolates from the urban lake in Brazil were identified as E. coli clone ST131 (Achtman scheme).25 Multilocus sequence typing performed on isolates from this cluster identified ST506 in the Pasteur scheme (previously linked to ST131 in the Achtman scheme27) and ST476 (first identified in this study). Two isolates from the urban lake in Brazil and one each from the sewage in the United States and the rural river in Brazil shared the same rep-PCR pattern, and all belonged to ST2 in the Pasteur scheme (akin to ST10 in the Achtman scheme), except one organism which was ST539, not sharing any alleles with ST2. Another cluster of E. coli isolates were identified as ST340.
Figure 4.
Dendrogram and band pattern generated by DiversiLab rep-PCR of Escherichia coli isolated from the study sites. Similarity ≥ 95% indicates clonally related strains. ESBL+ indicates a positive ESBL test by VITEK. “Sewer” designates isolates from raw sewage in Northeast Ohio, “DC” isolates from the urban lake in Brazil, and “Jenipapo” isolates from the rural river in Brazil. ESBL = extended-spectrum β-lactamases.
Among the 10 K. pneumoniae isolates, rep-PCR identified one cluster (> 95% similarity) that was defined as ST340 by MLST (using the Klebsiella typing scheme from the Pasteur institute).
Detection of AMR genes in water samples.
Because many bacterial species may serve as reservoirs for AMR genes, total DNA obtained from surface water samples was studied. We identified the carbapenemase gene blaOXA-48 in multiple samples of water from the Brazilian urban lake. This gene was found in eight of eight (100%) collection sites from the southern shore, and three of seven (43%) sites from the northern shore. In addition, blaKPC was present in samples from three sites and blaVIM-2 was discovered in samples from two sites in the southern shore. Of note, blaVIM-2 was also detected in the sample of raw sewage from Northeast Ohio. Plasmid-mediated quinolone resistance genes qnrS and aac(6')-lb-cr were detected in water samples from three and four sites at the Brazilian urban lake, respectively.
DISCUSSION
In this study, we sought to determine the presence of critically important contributors to AMR in surface waters in Brazil and their relationship with human fecal contamination. We found genes encoding clinically relevant carbapenemases (OXA-48, KPC, and VIM-2) and PMQR determinants (qnrS and aac(6′)-lb-cr) in water samples from Dique do Cabrito, a typical urban lake in Salvador, Brazil. Surface waters from this lake also had a high burden of other AMR phenotypes, including polymyxin-resistant Enterobacteriaceae, ESBL-producing K. pneumoniae, and ST131 E. coli associated with fluoroquinolone resistance and harboring CTX-M. Our analyses indicate a high degree of human fecal contamination, in survey sites with high percentages of resistant bacteria, especially in the northern half of the Brazilian lake (Figure 1). Indeed, the concentration of bacteria from human feces in one site in the urban lake in Brazil was like that of a sample of raw sewage from Northeast Ohio in the United States (Figure 2).
By contrast, AMR determinants were rare or not found in the study sites where human fecal bacteria were less frequent, such as a rural river in Brazil and an urban lake in Northeast Ohio. This was the case even though water samples from the rural river in Brazil contained ruminant fecal bacteria. Human fecal contamination, therefore, may contribute to the high burden of AMR determinants in surface water sampled from the urban lake in Brazil. Swimming and bathing occur at this site, with the ensuing risk of acquisition of AMR bacteria.
The presence of carbapenemase genes in samples from Dique do Cabrito is especially concerning, given the tremendous clinical import of carbapenem-resistant Gram-negative bacteria. Our screening, restricted to Enterobacteriaceae, did not isolate carbapenem-resistant bacteria in cultures from those same samples. This discrepancy may be explained by low sensitivity of phenotypic testing or poor genetic expression.28 Alternatively, carbapenemase genes in environmental samples may originate from other bacterial species. Although Enterobacteriaceae are the usual hosts of blaOXA-48 in clinical samples,29 the progenitor of blaOXA-48 in this instance may have been Shewanella spp., an environmental Gram-negative facultative aerobe that grows in fresh and salt water.30 The presence of blaVIM-2 in sewage from Northeast Ohio is also intriguing because bacteria harboring this carbapenemase are especially rare in the United States. One of the few reports of VIM-2–producing Pseudomonas aeruginosa in this country also originates from Northeast Ohio, and genetic analysis of that organism revealed that blaVIM-2 was contained in a mobile genetic element similar to those present in Salmonella.31 Interestingly, a cluster of blaVIM-2-containing P. aeruginosa of the same ST type (ST 233) was also found among residents of a skilled nursing facility in Chicago and previously in a hospital in the same city.32 Outside of the midwestern United States, VIM-producing P. aeruginosa was reported in Orlando, FL, as well.33
The detection of Enterobacteriaceae isolated from environmental water samples obtained from a lake surrounded by a presumably healthy community provides a wide-ranging sample of the “resistome” present in the fecal contents of thousands of residents. Therefore, our findings likely reflect the widespread presence of ESBL-producing Enterobacteriaceae in a Brazilian urban population. Reports originating from Brazil indicate high rates of ESBL production among clinical isolates of Enterobacteriaceae since the 2000s, up to 50% for K. pneumoniae and 17% for E. coli.34
The recovery of ST131 E. coli harboring blaCTX-M is also noteworthy because this is a successful global strain characterized by resistance to fluoroquinolones and cephalosporins with potentially increased virulence and deleterious clinical outcome.35 CTX-M is the dominant type of ESBL associated with fecal carriage in communities around the globe,4 and there is high carriage of ESBL-producing bacteria, often CTX-M–producing ST131 E. coli, in the intestinal tract of outpatients from high-, low-, and middle-income countries.36–38 The epidemiological success of this strain is likely due to antibiotic selection in humans and animals, compounded by human travel and wildlife migration, illustrating the “One Health” concept that links animal and human health. Of note, ST2 E. coli, reported in both nosocomial and agricultural CTX-M–possessing isolates, occurred in water samples from both locations in Brazil as well as the sewage sample from the United States.
Most ominous is the presence in the Brazilian lake of Enterobacteriaceae resistant to polymyxins. This class of antibiotics is reserved for the treatment of serious infections caused by extensively resistant Gram-negative bacteria in hospitalized patients. In Brazil, polymyxin resistance mediated by mcr-1 and mcr-2 has been reported in Enterobacteriaceae from environmental, animal, and human origin.39–41 Although these genes were not detected in isolates from this study, work is in progress to determine the genetic basis of this phenotype, including whole-genome sequencing of the isolates.
The dissemination of antibiotic-resistant bacteria in the environment including surface waters may explain the following paradox: by most measures, human consumption of antibiotics in Brazil is not significantly higher than in the United States,42 and yet, the rates of AMR are higher in Brazil.33 For instance, according to the Center for Disease Dynamics and Economic Policy (https://resistancemap.cddep.org/AntibioticUse.php), human consumption of fluoroquinolones in Brazil was 1,069 daily in defined doses (DDD)/1,000 population in 2015, which is similar to that of the United States (1,002 DDD/1,000 population). On the other hand, Brazil is the country with the third largest antimicrobial use in agriculture in the world (9%), but it is difficult to obtain data about the use of specific classes of antibiotics in animals.43 However, interest in complying with European market standards has led to raising antibiotic-free poultry in Brazil. In rural areas in the State of Bahia, we have solicited and not observed use in growth promotion, and cattle are primarily grass fed. In addition, non-prescription antibiotic use in both agriculture and medicine in Brazil was restricted in 2011.44 Although clandestine use is possible, evidence that non-prescription antimicrobial use is significantly different between the two countries is not available.45
This study is limited by the number of samples collected and by the cross-sectional rather than longitudinal nature of the collections. Also, the collection in Ohio was made in the fall, which is not the peak season for fecal contamination. Nevertheless, the magnitude of the differences observed between the Brazilian urban site and the rural site as well as Cleveland sewage sample indicate that qualitative differences are likely to remain. Our study only focused on determining contamination with human and ruminant feces; however, several other animals may serve as reservoirs of AMR such as migratory birds and companion animals, but their contributions were not studied. In addition, of the several genes that contribute to quinolone resistance, we focused on three common genes in Brazil and elsewhere, representing two mechanisms. For instance, the efflux pumps qepA and oqxAB were not queried; oqxAB has been linked with the use of the olaquindox and quinolones use as growth promoter, whereas qepA is an efflux pump with quinolones as substrate but of less clear epidemiologic importance in clinical isolates from Brazil.46 Neither did we query for qnrB, frequently detected among Enterobacteriaceae from poultry in the state of Sao Paulo in Brazil (> 20% of isolates). By contrast, the aac(6′)lb-cr gene is rare (< 1%) in poultry isolates but occurs in up to 40% of cases of nosocomial and community-acquired urinary tract infection.47,48 Because chromosomal mutations in the quinolone resistance determining region (QRDR) are necessary to obtain a fully quinolone-resistant phenotype, they are likely to be present along with PMQR in quinolone-resistant isolates from this study. Because QRDR mutations are not a transferable mechanism of resistance, our survey did not include them. Interestingly, PMQR genes qnrA, qnrB, and qnrS have been found in the chromosome of quinolone-resistant marine bacteria in aquaculture areas with heavy fluoroquinolone usage. Furthermore, there is commonality of quinolone resistance genes and class 1 integrons between marine bacteria and E. coli, suggesting horizontal gene transfer.49
Taken together, our observations suggest that surface waters contaminated because of poor sanitation may be a component of the “resistome.” This has been observed in high-income countries50 but may be especially important in low- and middle-income countries.51 Poor sanitation may promote the emergence, dissemination, and persistence of AMR even where selective pressure from human antibiotic consumption is not intense.6 The public health advantages of proper sanitation are well established. The relationships between surface water, sanitation, antibiotic exposure, bacterial gene transfer, and human colonization underscore the importance of determining effective “One Health” strategies to protect water resources and mitigate the global AMR crisis in human health.
Acknowledgments:
We wish to thank the Northeast Ohio Regional Sewer District for providing the sample of raw sewage and their useful suggestions. We also thank the team of curators of the Institut Pasteur MLST and Whole Genome MLST for curating their data and making it publicly available (http://bigsdp.pasteur.fr/).
REFERENCES
- 1.Woolhouse MEJ, Ward MJ, 2013. Microbiology. Sources of antimicrobial resistance. Science 341: 1460–1461. [DOI] [PubMed] [Google Scholar]
- 2.Vaz-Moreira I, Nunes OC, Manaia CM, 2014. Bacterial diversity and antibiotic resistance in water habitats: searching the links with the human microbiome. FEMS Microbiol Rev 38: 761–778. [DOI] [PubMed] [Google Scholar]
- 3.Szekeres E, Chiriac CM, Baricz A, Szőke-Nagy T, Lung I, Soran M-L, Rudi K, Dragos N, Coman C, 2018. Investigating antibiotics, antibiotic resistance genes, and microbial contaminants in groundwater in relation to the proximity of urban areas. Environ Pollut 236: 734–744. [DOI] [PubMed] [Google Scholar]
- 4.Woerther P-L, Burdet C, Chachaty E, Andremont A, 2013. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26: 744–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huijbers PM, Blaak H, de Jong MC, Graat EA, Vandenbroucke-Grauls CM, de Roda Husman AM, 2015. Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ Sci Technol 49: 11993–12004. [DOI] [PubMed] [Google Scholar]
- 6.Berglund B, 2015. Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. Infect Ecol Epidemiol 5: 28564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnold KE, Williams NJ, Bennett M, 2016. “Disperse abroad in the land”: the role of wildlife in the dissemination of antimicrobial resistance. Biol Lett 12: 20160137–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nellums LB, Thompson H, Holmes A, Castro-Sánchez E, Otter JA, Norredam M, Friedland JS, Hargreaves S, 2018. Antimicrobial resistance among migrants in Europe: a systematic review and meta-analysis. Lancet Infect Dis 18: 796–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arcilla MS, et al. 2017. Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect Dis 17: 78–85. [DOI] [PubMed] [Google Scholar]
- 10.Chan HL, Poon LM, Chan SG, Teo JW, 2011. The perils of medical tourism: NDM-1-positive Escherichia coli causing febrile neutropenia in a medical tourist. Singapore Med J 52: 299–302. [PubMed] [Google Scholar]
- 11.Baker S, 2015. Infectious disease. A return to the pre-antimicrobial era? Science 347: 1064–1066. [DOI] [PubMed] [Google Scholar]
- 12.Fletcher S, 2015. Understanding the contribution of environmental factors in the spread of antimicrobial resistance. Environ Health Prev Med 20: 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pruden A, 2014. Balancing water sustainability and public health goals in the face of growing concerns about antibiotic resistance. Environ Sci Technol 48: 5–14. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization , 2017. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery and Development of New Antibiotics. Available at: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/. Accessed March 21, 2018. [Google Scholar]
- 15.Tacconelli E, et al. WHO Pathogens Priority List Working Group , 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18: 318–327. [DOI] [PubMed] [Google Scholar]
- 16.Hadjadj L, Riziki T, Zhu Y, Li J, Diene SM, Rolain J-M, 2017. Study of mcr-1 gene-mediated colistin resistance in Enterobacteriaceae isolated from humans and animals in different countries. Genes (Basel) 8: E394–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbosa L, Silva L, Reis E, Azevedo T, Costa J, Blank W, Reis M, Blanton R, 2013. Characteristics of the human host have little influence on which local Schistosoma mansoni populations are acquired. PLoS Negl Trop Dis 7: e2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanton RE, et al. 2015. The relative contribution of immigration or local increase for persistence of urban schistosomiasis in Salvador, Bahia, Brazil. PLoS Negl Trop Dis 9: e0003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okeke BC, Thomson MS, Moss EM, 2011. Occurrence, molecular characterization and antibiogram of water quality indicator bacteria in river water serving a water treatment plant. Sci Total Environ 409: 4979–4985. [DOI] [PubMed] [Google Scholar]
- 20.Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A, 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38: 1–40. [DOI] [PubMed] [Google Scholar]
- 21.Ponce-Terashima R, Koskey AM, Reis MG, McLellan SL, Blanton RE, 2014. Sources and distribution of surface water fecal contamination and prevalence of schistosomiasis in a Brazilian village. PLoS Negl Trop Dis 8: e3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojas LJ, et al. 2017. Antibacterial resistance leadership group. Colistin resistance in carbapenem-resistant Klebsiella pneumoniae: laboratory detection and impact on mortality. Clin Infect Dis 64: 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wintermans BB, Reuland EA, Wintermans RG, Bergmans AM, Kluytmans JA, 2013. The cost-effectiveness of ESBL detection: towards molecular detection methods? Clin Microbiol Infect 19: 662–665. [DOI] [PubMed] [Google Scholar]
- 24.Endimiani A, et al. 2009. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the eastern USA. J Antimicrob Chemother 63: 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitout JD, Campbell L, Church DL, Wang PW, Guttman DS, Gregson DB, 2009. Using a commercial DiversiLab semiautomated repetitive sequence-based PCR typing technique for identification of Escherichia coli clone ST131 producing CTX-M-15. J Clin Microbiol 47: 1212–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magiorakos AP, et al. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18: 268–281. [DOI] [PubMed] [Google Scholar]
- 27.Johnson J, Clermont O, Johnston B, Clabots C, Tchesnokova V, Sokurenko E, Junka A, Maczynska B, Denamur E, 2014. Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131. J Clin Microbiol 52: 1358–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nordmann P, Poirel L, 2013. Strategies for identification of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 68: 487–489. [DOI] [PubMed] [Google Scholar]
- 29.Patel G, Bonomo RA, 2013. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol 4: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poirel L, Héritier C, Nordmann P, 2004. Chromosome-encoded ambler class D beta-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase. Antimicrob Agents Chemother 48: 348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez F, et al. 2014. Extensively drug-resistant Pseudomonas aeruginosa isolates containing blaVIM-2 and elements of Salmonella genomic island 2: a new genetic resistance determinant in northeast Ohio. Antimicrob Agents Chemother 58: 5929–5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clegg WJ, et al. 2018. Notes from the field: large cluster of Verona integron-encoded metallo-beta-lactamase–producing carbapenem-resistant Pseudomonas aeruginosa isolates colonizing residents at a skilled nursing facility—Chicago, Illinois, November 2016–March 2018. Morb Mortal Wkly Rep 67: 1130–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rankin D, Caicedo L, Dotson N, Gable P, Chu A, 2018. Notes from the field: Verona integron-encoded metallo-beta-lactamase-producing Pseudomonas aeruginosa outbreak in a long-term acute care hospital–Orange County, Florida, 2017. MMWR Morb Mortal Wkly Rep 67: 611–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gales AC, Castanheira M, Jones RN, Sader HS, 2012. Antimicrobial resistance among Gram-negative bacilli isolated from Latin America: results from SENTRY Antimicrobial Surveillance Program (Latin America, 2008–2010). Diagn Microbiol Infect Dis 73: 354–360. [DOI] [PubMed] [Google Scholar]
- 35.Nicolas-Chanoine MH, Bertrand X, Madec JY, 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27: 543–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruppé E, et al. 2015. High rate of acquisition but short duration of carriage of multidrug-resistant Enterobacteriaceae after travel to the tropics. Clin Infect Dis 61: 593–600. [DOI] [PubMed] [Google Scholar]
- 37.Birgy A, Cohen R, Levy C, Bidet P, Courroux C, Benani M, Thollot F, Bingen E, 2012. Community faecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae in French children. BMC Infect Dis 12: 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hijazi SM, Fawzi MA, Ali FM, Abd El Galil KH, 2016. Prevalence and characterization of extended-spectrum beta-lactamases producing Enterobacteriaceae in healthy children and associated risk factors. Ann Clin Microbiol Antimicrob 15: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandes MR, Sellera FP, Esposito F, Sabino CP, Cerdeira L, Lincopan N, 2017. Colistin-resistant mcr-1-positive Escherichia coli on public beaches, an infectious threat emerging in recreational waters. Antimicrob Agents Chemother 61: e00234-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aires CAM, da Conceição-Neto OC, Tavares E Oliveira TR, Dias CF, Montezzi LF, Picão RC, Albano RM, Asensi MD, Carvalho-Assef APD, 2017. Emergence of the plasmid-mediated mcr-1 gene in clinical KPC-2-producing Klebsiella pneumoniae sequence type 392 in Brazil. Antimicrob Agents Chemother 61: e00317-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monte DF, Mem A, Fernandes MR, Cerdeira L, Esposito F, Galvão JA, Franco BDGM, Lincopan N, Landgraf M, 2017. Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob Agents Chemother 61: e02718-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gelband H, Miller-Petrie M, Pant S, Gandra S, Levinson J, Barter D, White A, Laxminarayan R, 2015. The state of the world’s antibiotics. Wound Heal South Afr 8: 30–34. [Google Scholar]
- 43.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R, 2015. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA 112: 5649–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maron DF, Smith TJ, Nachman KE, 2013. Restrictions on antimicrobial use in food animal production: an international regulatory and economic survey. Glob Health 9: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morgan DJ, Okeke IN, Laxminarayan R, Perencevich EN, Weisenberg S, 2011. Non-prescription antimicrobial use worldwide: a systematic review. Lancet Infect Dis 11: 692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Viana AL, Cayô R, Avelino CC, Gales AC, Franco MC, Minarini LA, 2013. Extended-spectrum β-lactamases in Enterobacteriaceae isolated in Brazil carry distinct types of plasmid-mediated quinolone resistance genes. J Med Microbiol 62: 1326–1331. [DOI] [PubMed] [Google Scholar]
- 47.Volcão LM, et al. 2018. High frequency of aac(6′)-Ib-cr gene associated with double mutations in gyrA and parC in Escherichia coli isolates from patients with urinary tract infections. J Glob Antimicrob Resist 13: 180–183. [DOI] [PubMed] [Google Scholar]
- 48.Ferreira JC, Penha Filho RAC, Kuaye APY, Andrade LN, Berchieri Junior A, Darini ALDC, 2018. Identification and characterization of plasmid-mediated quinolone resistance determinants in Enterobacteriaceae isolated from healthy poultry in Brazil. Infect Genet Evol 60: 66–70. [DOI] [PubMed] [Google Scholar]
- 49.Tomova A, Ivanova L, Buschmann AH, Godfrey HP, Cabello FC, 2018. Plasmid-mediated quinolone resistance (PMQR) genes and class 1 integrons in quinolone-resistant marine bacteria and clinical isolates of Escherichia coli from an aquacultural area. Microb Ecol 75: 104–112. [DOI] [PubMed] [Google Scholar]
- 50.Zurfluh K, Hächler H, Nüesch-Inderbinen M, Stephan R, 2013. Characteristics of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae isolates from rivers and lakes in Switzerland. Appl Environ Microbiol 79: 3021–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alouache S, Estepa V, Messai Y, Ruiz E, Torres C, Bakour R, 2014. Characterization of ESBLs and associated quinolone resistance in Escherichia coli and Klebsiella pneumoniae isolates from an urban wastewater treatment plant in Algeria. Microb Drug Resist 20: 30–38. [DOI] [PubMed] [Google Scholar]
- 52.Bernhard AE, Field KG, 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl Environ Microbiol 66: 4571–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JD, 2012. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol 50: 3877–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daoud Z, Salem Sokhn E, Masri K, Matar GM, Doron S, 2015. Escherichia coli isolated from urinary tract infections of Lebanese patients between 2005 and 2012: epidemiology and profiles of resistance. Front Med 2: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guillard T, Cavallo JD, Cambau E, Duval V, Bajolet O, Brasme L, de Champs C, Vernet-Garnier V, 2010. Real-time PCR for fast detection of plasmid-mediated qnr genes in extended spectrum beta-lactamase producing Enterobacteriaceae. Pathol Biol (Paris) 58: 430–433. [DOI] [PubMed] [Google Scholar]
- 56.Pereira AS, Andrade SS, Monteiro J, Sader HS, Pignatari AC, Gales AC, 2007. Evaluation of the susceptibility profiles, genetic similarity and presence of qnr gene in Escherichia coli resistant to ciprofloxacin isolated in Brazilian hospitals. Braz J Infect Dis 11: 40–43. [DOI] [PubMed] [Google Scholar]
- 57.Yu T, Jiang X, Fu K, Liu B, Xu D, Ji S, Zhou L, 2015. Detection of extended-spectrum β-lactamase and plasmid-mediated quinolone resistance determinants in Escherichia coli isolates from retail meat in China. J Food Sci 80: M1039–M1043. [DOI] [PubMed] [Google Scholar]