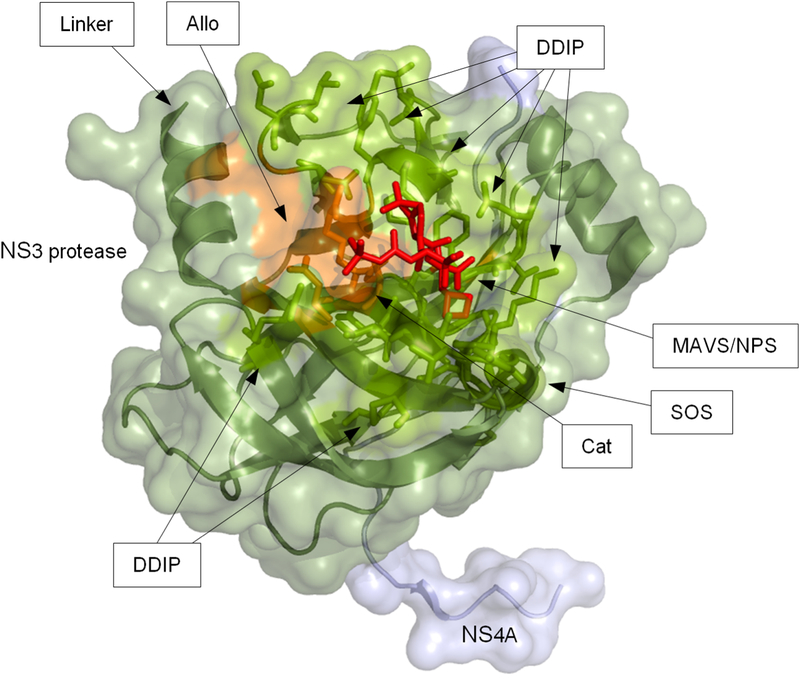
Fig 1. 3D protein structure of the NS3–4A protease domain.
The protein structure shows a subtype 1a NS3 protease domain from Protein Data Bank entry 2OC8 co-crystallized with NS4A (light blue) and a linear protease inhibitor (PI) (purple stick model). The protein backbone is given as ribbon model with transparent surface depiction. Residues where PI resistance-associated amino acid substitutions occur are highlighted as orange stick models; functionally important residues are shown as green stick models. Functionally important sites are indicated by arrows: allosteric site (Allo); protease active/catalytic site (Cat); domain-domain interaction sites (DDIP); linker region (Linker); MAVS binding site (MAVS); binding site for the natural protease substrate (NPS); SOS-binding site for TRIF (SOS).