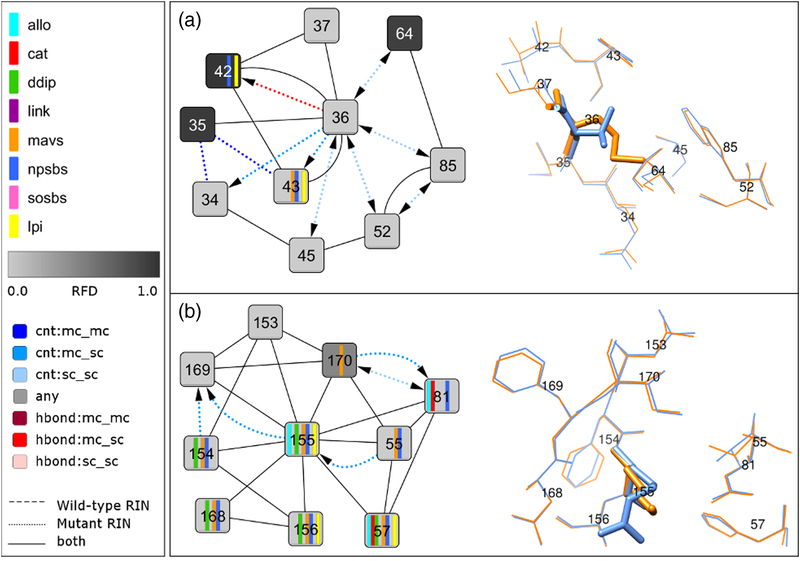
Fig 4. Network comparison view between wild-type and mutant structure for V36M and R155K and corresponding 3D structure close-up.
(A) The comparison network view between the subtype 1a consensus residue-interaction network and the residue-interaction network of the V36M mutant structure from Protein Data Bank entry 2QV1 focuses on the direct noncovalent interactions of residue 36 with its neighbors. The corresponding residues in the structure are shown as blue and orange sticks, respectively. (B) The comparison network view between the subtype 1a consensus residue-interaction network and the residue-interaction network of the R155K mutant structure from Protein Data Bank entry 2OIN focuses on the direct noncovalent interactions of residue 155 with its neighbors. The corresponding residues in the structure are shown as blue and orange sticks, respectively.