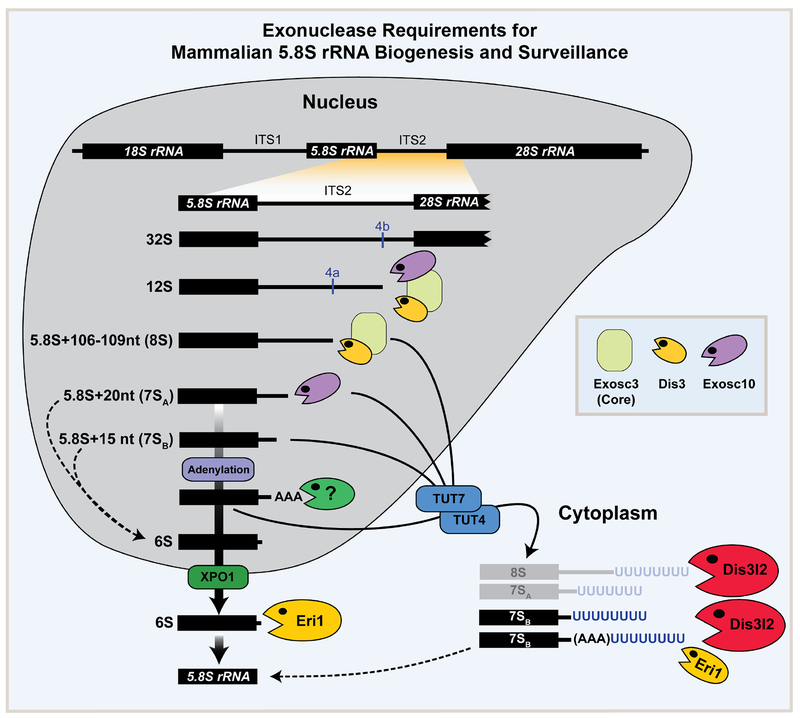
Fig. 6. Exonuclease requirements for mammalian 5.8S rRNA biogenesis and surveillance.
A model summarizing the function of various exoribonucleases in the processing of 5.8S rRNA intermediates. During step-wise maturation of 5.8S rRNA, exosome and Exosc10 are responsible for processing of the 8S rRNA and 7SA (5.8S+20nt) intermediates, respectively. 7SB rRNA (5.8S+15nt) which is a potential product of Exosc10 can be adenylated (by an unknown enzyme) and/or further processed by an unknown nuclease to the 6S pre-rRNA that processed by Eri1 to mature 5.8S rRNA in the cytoplasm (thick solid arrows). Failure of any of these processing steps results in oligouridylation by TUT4/7 and processing by Dis3l2 and Eri1 in the cytoplasm (arrows). Dashed lines represent possible alternative steps in the 5.8S rRNA processing pathway.