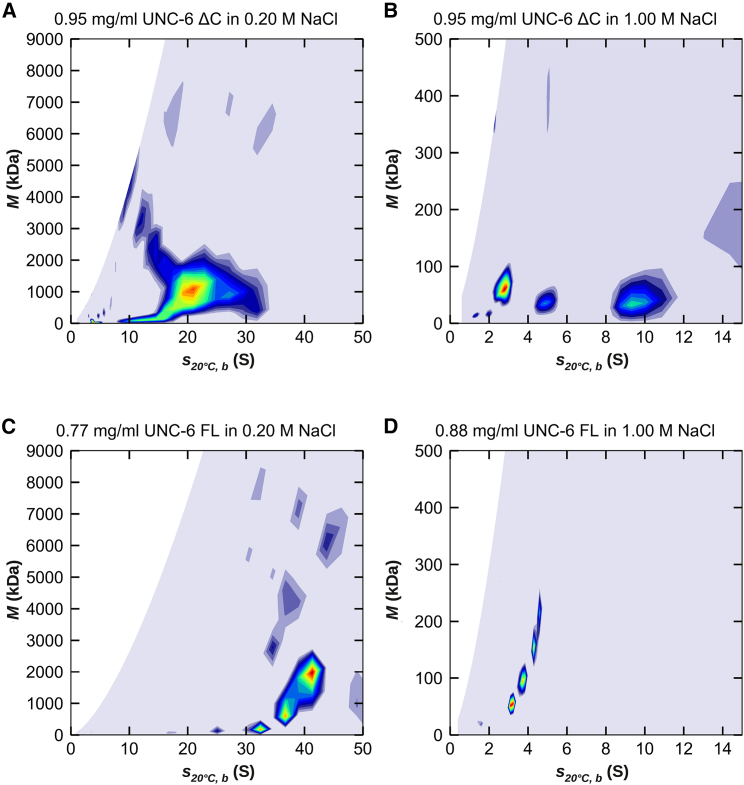
Figure 3.
Two-dimensional c(s, M) distributions obtained from the interference data of (A) C. elegans UNC-6 ΔC in low-ionic strength buffer, (B) C. elegans UNC-6 ΔC in high-ionic strength buffer, (C) C. elegans UNC-6 FL in low-ionic strength buffer, and (D) C. elegans UNC-6 FL in high-ionic strength buffer at concentrations close to 1 mg/mL. In low-ionic strength, the protein forms assemblies of high molecular mass in the range of 1–2 MDa. At high-ionic strength, the formation of these assemblies is suppressed, and the distributions suggest populations of monomers, dimers, and multimers in the case of the ΔC truncation and a monomer/dimer equilibrium (plus a small amount of a larger species) in the case of the FL version. Interestingly, in low-ionic strength, the assemblies of the FL protein have sedimentation coefficients twice as large as the truncated protein; however, their masses are similar. This suggests that the shapes of the assemblies differ between the two protein versions. To see this figure in color, go online.