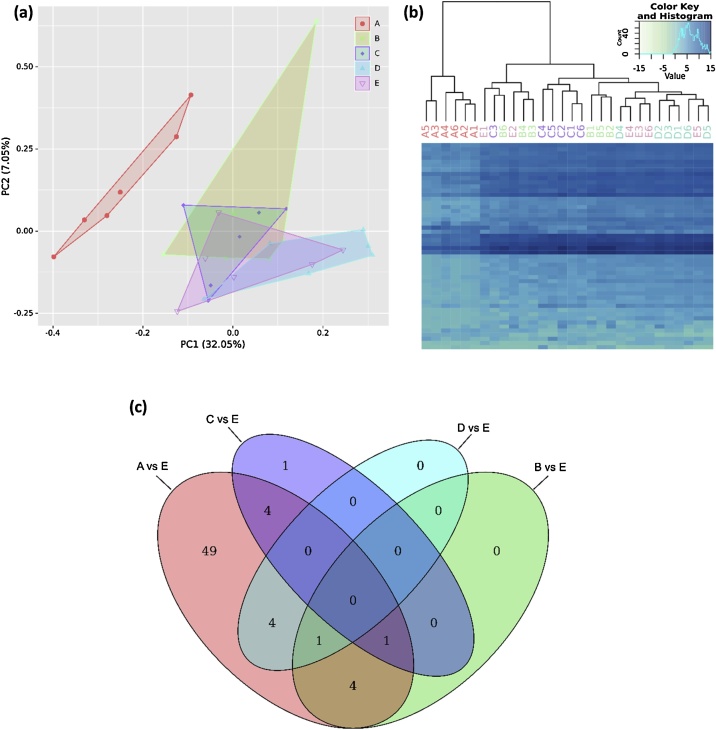
Fig. 5.
Exploratory data analysis of differences in urinary miRNAs derived from distinct uEV purification strategies (n = 6): spin column chromatography (A), immunoaffinity (B), precipitation (C), membrane affinity (D) and ultracentrifugation combined with density gradient (E). (a) Principal component analysis of the complete miRNA gene set showing the first and second principal component (PC) of explained variance. (b) Heatmap and hierarchical cluster analysis indicated strong separation of A from other methods. (c) Venn diagram of significantly differentially detected miRNAs compared to E. Results were determined by DESeq2-based differential gene expression analysis and filtered for adjusted p value < 0.05 and log2 fold change > |1|.