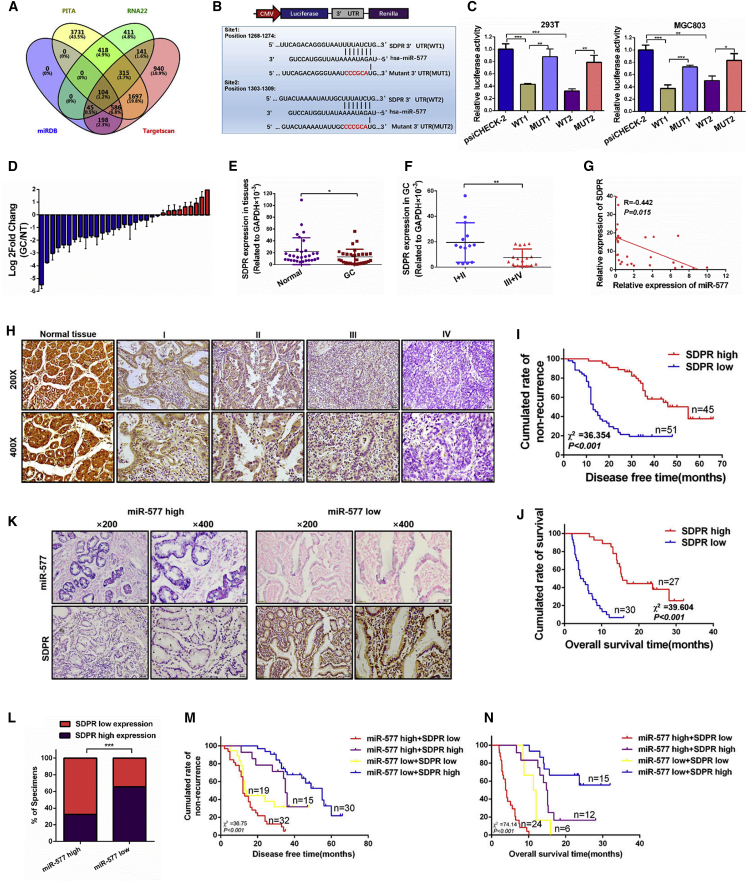
Figure 5.
SDPR Is a Metastasis Suppressor Downstream of miR-577 in GC
(A) Bioinformatics prediction of potential miR-577 targets by four common miRNA databases (TargetScan, PITA, RNA22, and miRDB). (B) Potential miR-577 binding sequences in the 3′ UTR of SDPR mRNAs and the structure of the WT (WT1 and WT2) and mutant (Mut1 and Mut2) luciferase reporters. (C) Relative luciferase activity of the indicated 3′UTR vectors in 293T and MGC803 cells co-transfected with miR-577 AgomiR and SDPR plasmids. (D) qRT-PCR analysis of SDPR expression in 30 pairs of GC specimens and normal tissues, which are normalized to an endogenous GAPDH RNA control. Data represent the mean ± SD. (E and F) The SDPR expression in 30 pairs of GC specimens and normal tissues (E) and in GC tissues in the TNM stages I and II or stages III and IV (F). *p < 0.05; **p < 0.01. (G) Correlation between miR-577 and SDPR mRNA expression levels. Two-tailed Spearman’s correlation analysis. Data represent mean ± SD; p = 0.015. (H) Immunohistochemistry (IHC) analysis of SDPR expression in 153 human normal gastric tissues and GC specimens from TNM stage I–IV patients. (I and J) Retrospective analysis of Kaplan-Meier plots for SDPR expression and the association with disease-free survival (I) and overall survival (J). ***p < 0.001. (K and L) Representative images of successive IHC and ISH staining of miR-577 and SDPR expression in GC tumor specimens (K). Bars represent the percentage of specimens (L). ***p < 0.001. (M and N) Retrospective analysis of Kaplan-Meier plots of the co-expression of SDPR and miR-577 in association with disease-free survival (M) and overall survival (N). ***p < 0.001.