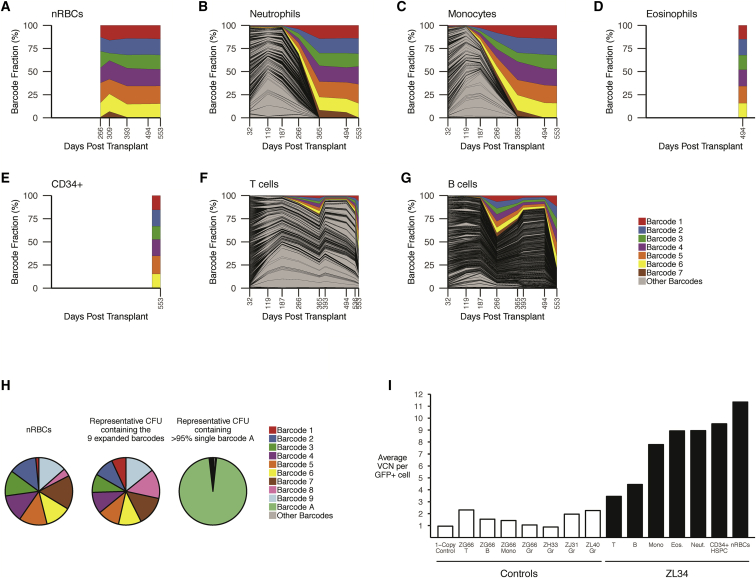
Figure 2.
Clonal Tracking via Barcode Retrieval in ZL34
(A–G) Longitudinal clonal tracking of barcodes retrieved from nRBCs (A), neutrophils (B), monocytes (C), eosinophils (D), BM CD34+ cells (E), T cells (F), and B cells (G). Each graph shows the fractional contributions from individual barcodes over time, with each colored ribbon representing the expanded barcodes detected via standard barcode recovery and all other barcodes shaded in gray. (H) Modified barcode recovery using the alternative forward primer on ZL34 nRBCs (393 days post-transplant), showing retrieval of 9 expanded barcodes. Modified barcode recovery using the same primers was performed on single-cell-derived GFP+ CFUs from ZL34 BM CD34+ cells (266 days post-transplant) plated at low density. Representative barcode analysis on a myeloid CFU containing all 9 barcodes is shown, matching those found in the circulating nRBC, confirming integration of the 9 barcodes in one original HSPC. For comparison, barcode analysis on a myeloid CFU from the same plate found to contain a single barcode, barcode A, is shown, confirming lack of cellular contamination across the plate that would complicate analysis. The small sector shown in gray represents multiple other barcodes with very low read counts, likely resulting from residual single cells not forming CFUs. (I) Vector copy number per GFP+ cell quantified via qPCR on Gr (granulocytes), Mono (monocytes), T cells, B cells, Eos (eosinophils), nRBCs, and CD34+ BM HSPCs from ZL34 (black bars, days 494–553 post-transplantation) and additional barcoded transplanted macaques (white bars, days 278–1,085 post-transplantation).