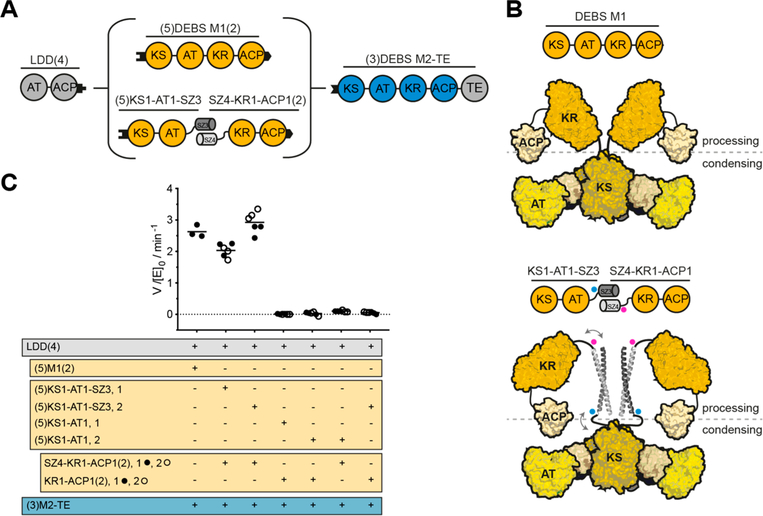
Figure 2.
Use of SYNZIP domains in the design of catalytically efficient PKSs harboring a split module. (A) Design of a bimodular DEBS derivative comprised of LDD(4), intact (5)M1(2) or a split version thereof, and (3)M2-TE. (B) Model of DEBS M1 with the natural AT-KR linker (top) and the SYNZIP-containing variant (bottom). This model was built based on SAXS analysis of DEBS M3.32 Fusion sites of the SYNZIP domains to either the AT or KR are indicated by blue and pink dots, illustrating the parallel orientation of the heterospecific coiled-coil. An eight-residue flexible Gly-Ser linker is used to connect the SYNZIP domain to the PKS protein (see protein sequence Table S2). (C) Turnover rates of bimodular PKSs employing M1 or a split M1. Except for intact M1, at least two independently purified protein preparations were evaluated (1 or 2): each preparation of the N-terminal part is shown in a separate column, whereas preparations of the C-terminal part are indicated as black and white dots. All initial rate data was obtained at 2 μM enzyme concentration and nonlimiting concentrations of propionyl-CoA, methylmalonyl-CoA, and NADPH. Measurements were performed in triplicate, and the grand mean is indicated.