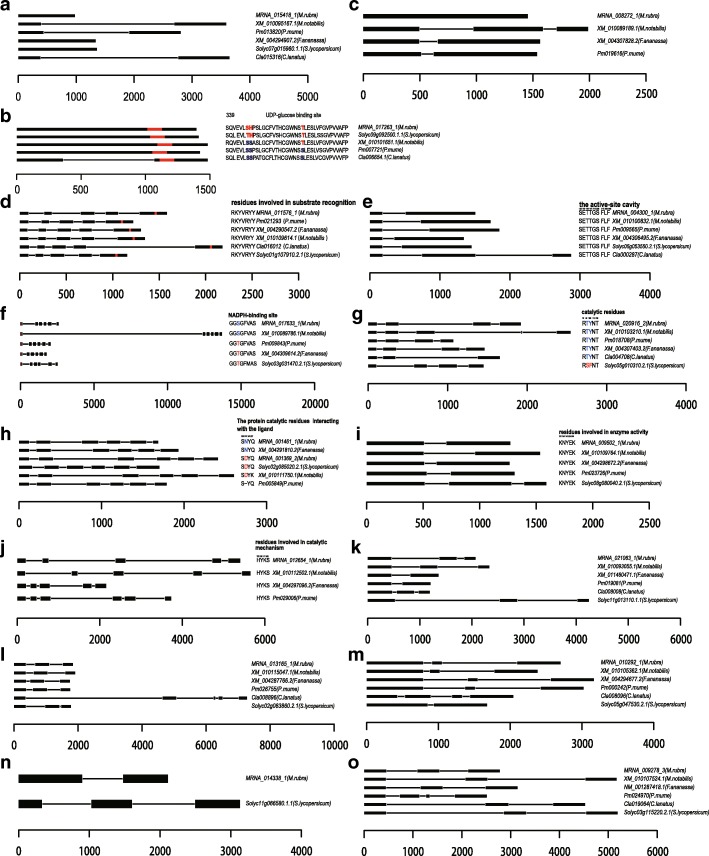
Fig. 5.
Gene structure and/or single amino acid changes in functionally active residues for gene expansions in flavonoid and anthocyanin biosynthesis pathways. (a) shikimate O-hydroxycinnamoyltransferase (HCT, K13065). (b) anthocyanidin 3-O-glucoside 5-O-glucosyltransferase (UGT75C1, K12338). (c) anthocyanidin 3-O-glucosyltransferase (BZ1, K12930). (d) Caffeoyl-CoA O-methyltransferase (CCoAOMT, K00588). (e) chalcone synthase (CHS, K00660). (f) anthocyanidin reductase (ANR, K08695). (g) chalcone isomerase (CHI, K01859). (h) bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase (DFR, K13082). (i) leucoanthocyanidin dioxygenase (LDOX, K05277). (j) leucoanthocyanidin reductase (LAR, K13081). (k) flavonol synthase (FLS, K05278). (l) naringenin 3-dioxygenase (K00475). (m) trans-cinnamate 4-monooxygenase (CYP73A, K00487). (n) flavonoid 3',5'-hydroxylase (K13083). (o) flavonoid 3'-monooxygenase (K05280)