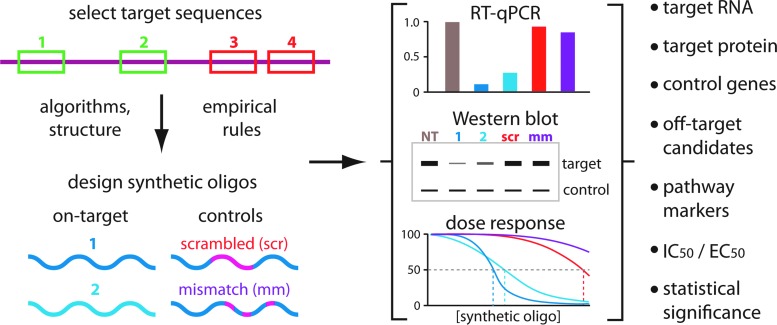
FIG. 3.
General guidelines for performing convincing gene expression knockdown experiments with synthetic oligonucleotides. Once a specific target RNA has been identified, several putative target sequences are chosen using empirical rules (such as preferred target regions, desired experimental outcome, and chemical modification limitations), algorithms to predict the best sites (with minimal off targets), and available structural information (to improve accessibility). At least two on-target oligonucleotides and two control oligonucleotides should be designed. Controls should be at least one scrambled and one mismatch oligonucleotide based on the lead on-target oligonucleotide. Titrations should be performed to obtain dose–response curves. RNA levels and protein levels (when applicable) should be measured. EC50, effective concentration; IC50, inhibitory concentration; RT-qPCR, reverse transcription quantitative PCR.