FIGURE 4.
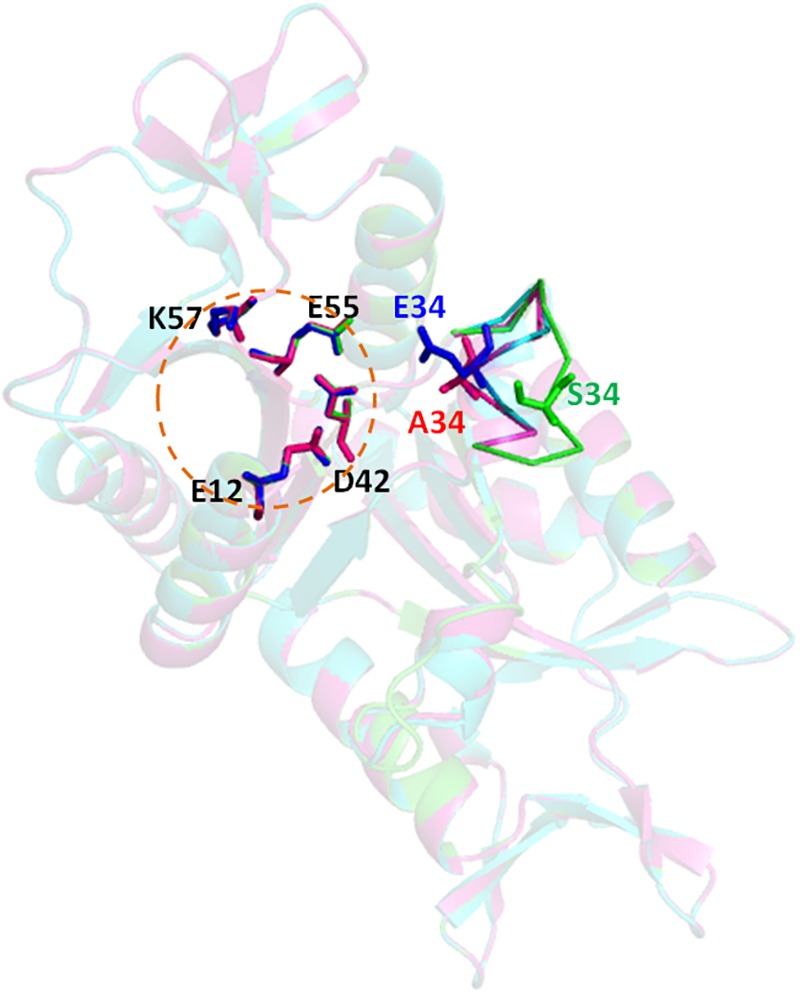
Structure modeling of Hjc, S34A, and S34E. The structural model were built using the online server SWISS-MODEL (https://swissmodel.expasy.org/) based on the S. solfataricus Hjc structure (PDB: 1HH1) and analyzed by PyMOL. The three structures (dimer) are aligned together in which wild type (S34), S34A, and S34E are in blue, pink, and green colors, respectively. The circular dotted line indicates the catalytic center consisting of four highly conserved residues, E12, D42, E55, and K57. A five-amino acid loop containing the mutated residue is shown as sticks. The distances between the 34th amino acid to the closest catalytic residue D42 are D42-S34∼9.9Å, D42-A34∼5.4Å, and D42-E34∼4.7Å, respectively.
