Figure 7. GABAergic Neurons Exhibit Greater Molecular Sex Differences than Glutamatergic Neurons.
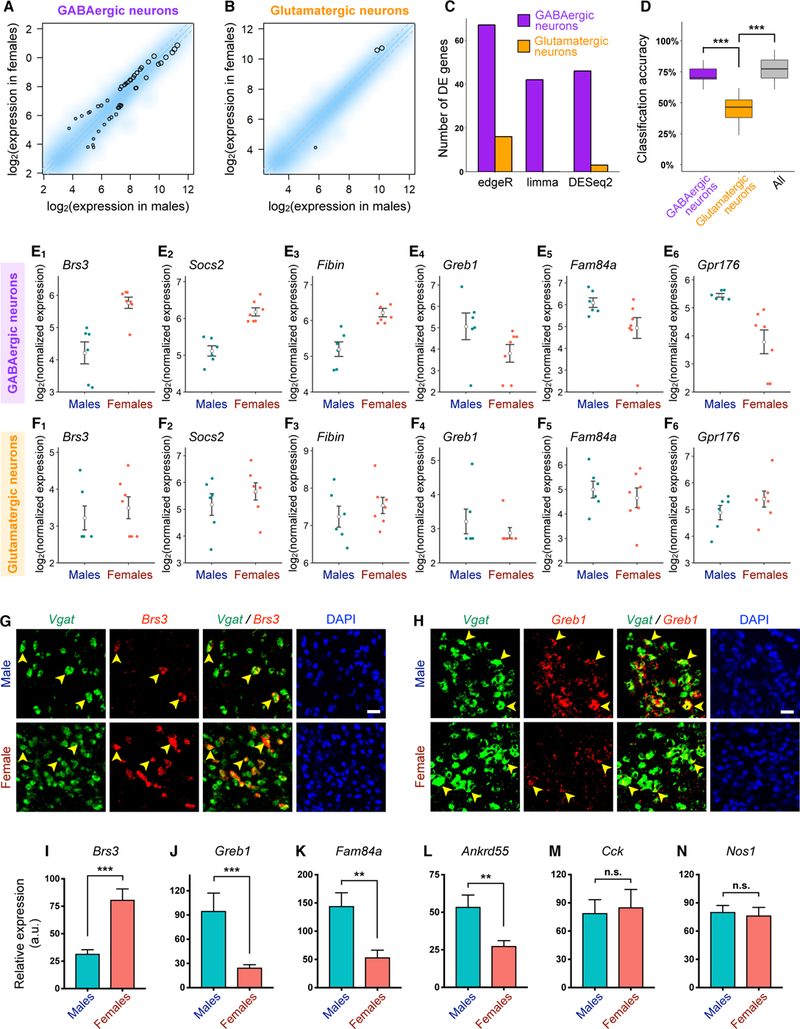
(A-B) Smoothed scatter plots showing expression of neuronally enriched genes in females versus males in GABAergic (A) and glutamatergic (B) neurons. Expression level was normalized for each sample (STAR Methods) and averaged across male and female samples. Color reflects density of genes. Black circles highlight genes significantly differentially expressed between males and females based on the DESeq2 method (STAR Methods). Circle size is proportional to mean expression level across all samples. Grey dotted lines = fold change cutoff of 1.3.
(C) Number of significantly differentially expressed genes in GABAergic and glutamatergic neurons using three different approaches (STAR Methods).
(D) Box plots showing accuracy of classification of male versus female samples using linear discriminative analysis of GABAergic, glutamatergic, or all neurons.
(E-F) Dot plots showing expression level of representative genes with significantly higher (E1-E3) or lower (E4-E6) expression in females versus males in GABAergic neurons (E) but not in glutamatergic neurons (F). Each dot represents the summed expression level of each gene over all GABAergic or glutamatergic neurons in an independent biological sample (n = 6 male samples (12 mice) and 7 female samples (20 mice; STAR Methods). Open circles and error bars: mean ± SEM.
(G-H) Representative double FISH images showing sex differences in the expression of Brs3 (G) and Greb1 (H) in Vgat+ cells (arrowheads) in the MeApd. Scale bar = 50 µm.
(I-N) Quantification of fluorescence intensity of probes specific for each transcript in Vgat+ cells in sections of male and female MeApd. n = 5–9 different sections from 3–5 mice for each sex (STAR Methods).
Mean ± SEM; Wilcoxon rank-sum test. n.s. not significant, **p < 0.01, ***p < 0.001.
