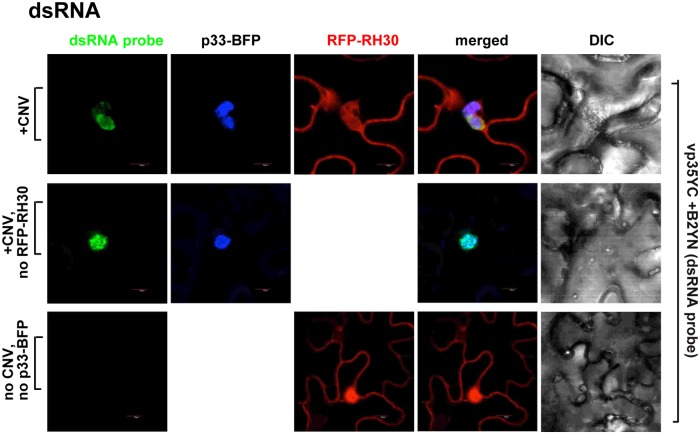
Fig 10. Co-localization of the viral double-stranded gRNA with RH30 in whole plants infected with CNV.
The CNV genomic dsRNA replication intermediate was detected via a dsRNA detector assay based on dsRNA binding-dependent fluorescence complementation assay [69]. The assay was performed with two dsRNA binding proteins (i.e., vp35 and B2), which are fused to N- and C-terminal halves of the yellow fluorescence protein (YFP), respectively. Simultaneous binding of the two fusion proteins to the same CNV dsRNA replication intermediate leads to the restoration of YFP fluorescence, allowing the visualization of the viral dsRNA replication intermediate location via confocal microscopy. The dsRNA sensor B2YN and VP35YC plasmids were agro-infiltrated into N. benthamiana leaves at OD600 of 0.15, respectively, together with RFP-RH30 and p33-BFP at OD600 of 0.5. CNV infection was initiated via agro-infiltration (OD600 of 0.15). Leaves were harvested and then immediately subjected to confocal microscopic analysis 2 days after agro-infiltration. The fluorescence complementation was detected via the GFP channel (excitation/emission: 488nm/500-530nm). Top panel: viral dsRNA replication intermediate is co-localized with RFP-RH30 within the replication compartment, which is marked by TBSV p33-BFP. Middle panel: no expression of RFP-RH30 was used as control. Bottom panel: N. benthamiana leaves with no viral components expressed were used as control. Expression of the above proteins from 35S promoter was done after co-agroinfiltration into N. benthamiana leaves. Scale bars represent 10 μm. Each experiment was repeated.