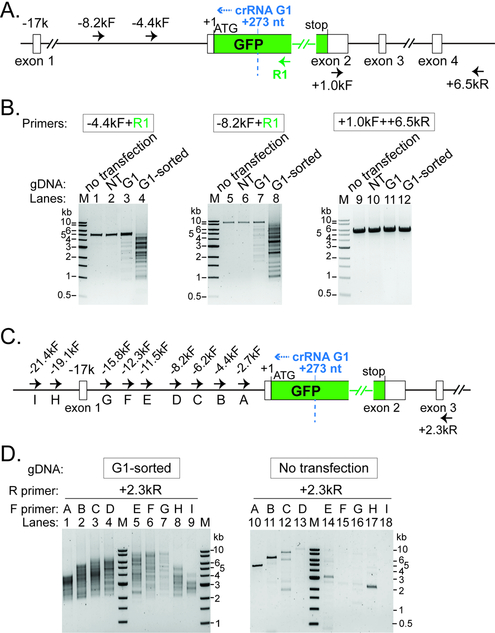
Figure 3. Long-range PCR based characterization of genomic lesions induced by Type I CRISPR-Cas.
(A) (C) Schematic of the DNMT3B-EGFP locus and annealing sites for five PCR primers used in (B) or ten PCR primers used in (D). Positions relative to the EGFP translation start site (+1) are indicated. Recognition sites (2nd nt of the PAM) for Cascade- G1 are marked by the dashed blue line. Blue arrowhead, direction of Cas3 translocation. (B) Characterization of genomic lesions by long-range PCRs. A collection of DNA lesions was introduced upstream of GFP by a GFP-targeting Cascade-G1 and Cas3, in the sorted EGFP-negative/tdTomato-positive population, as well as in unsorted total cells. NT, total cells treated with a non-targeting Cascade and Cas3. PCR primers used are indicated and their annealing sites depicted in (A). (D) Heterogeneous large genomic deletions were introduced by Cascade-G1/Cas3 in the ~20 kb region upstream of GFP, revealed by serial long-range PCRs with tiling forward primers. NT, non-targeting Cascade. PCR primers used are indicated and their annealing sites depicted in (C). M, DNA size markers.