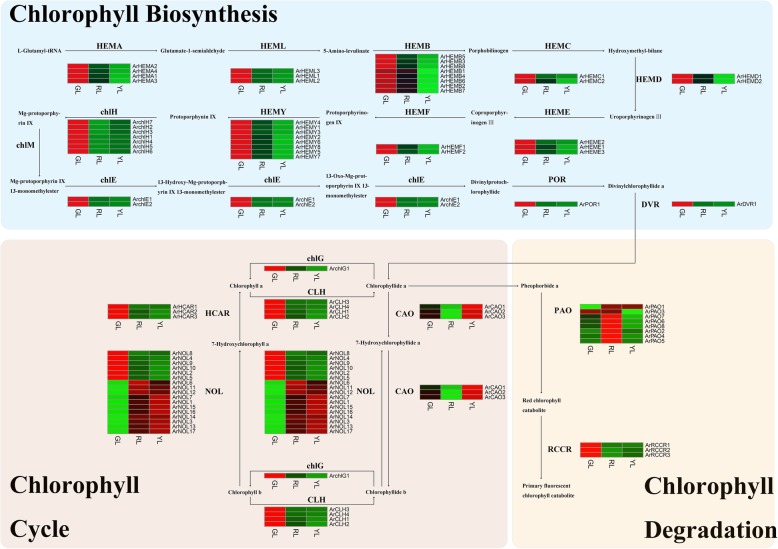
Fig. 5.
Heatmap of differentially expressed genes (DEGs) related to chlorophyll metabolism. GL, green leaves; RL, red leaves; YL, yellow leaves; HEMA, glumly-tRNA reductase; HEML, Glutamate-1-semialdehyde 2,1-aminomutase; HEMB, Glutamate-1-semialdehyde 2,1-aminomutase; HEMC, Glutamate-1-semialdehyde 2,1-aminomutase; HEMD, Uroporphyrinogen-III synthase; HEME, Uroporphyrinogen decarboxylase; HEMF, Coproporphyrinogen III oxidase; HEMY, Oxygen-dependent protoporphyrinogen oxidase; chlH, Magnesium chelatase subunit H; chlM, Magnesium-protoporphyrin IX methyltransferase; chlE, Magnesium-protoporphyrin IX methyltransferase; POR, protochlorophyllide reductase; DVR, divinyl chlorophyllide a 8-vinyl-reductase; CAO, chlorophyllide a oxygenase; chlG, chlorophyll synthase; CLH, chlorophyllase; HCAR, 7-hydroxymethyl chlorophyll-a reductase; NOL, non-yellow coloring-like; PaO, pheophorbide a oxygenase; RCCR, red chlorophyll catabolite reductase