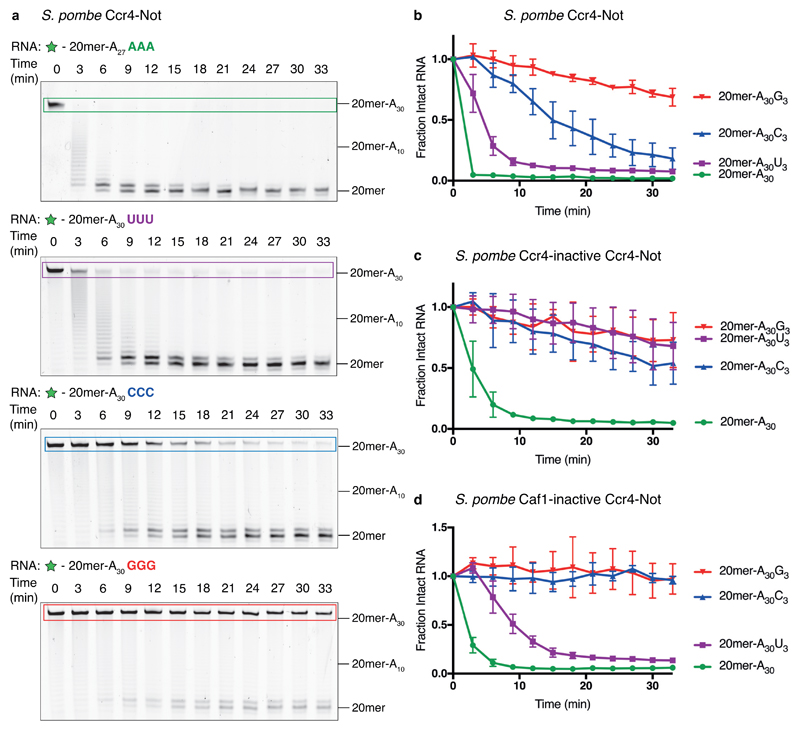
Figure 2. Ccr4–Not is inhibited by 3′ guanosines.
a, Denaturing RNA gels showing deadenylation by recombinant S. pombe Ccr4–Not on 5′ 6-FAM-labeled (green star) RNA substrates consisting of a 20mer non-poly(A) sequence (See Fig. 1a) followed by 30 adenosines. Where indicated, the substrate contains three additional non-A nucleotides at the 3′ end. These gels are representative of identical experiments performed 3 times. Uncropped gel images are shown in Supplementary Data Set 1. b-d, Analysis of deadenylation on poly(A) substrates with different 3′ nucleotides. Disappearance of the intact substrate was quantified by densitometry of the fluorescently labeled, full-length RNA. Data points were normalized to time = 0, and are connected by straight lines for clarity. Assays were carried out in triplicate (n = 3 independent experiments), the data points shown represent the mean, and error bars represent standard deviation. Assays are shown for wild-type S. pombe Ccr4–Not (b), Ccr4-inactive Ccr4–Not (c) and Caf1-inactive Ccr4–Not (d).