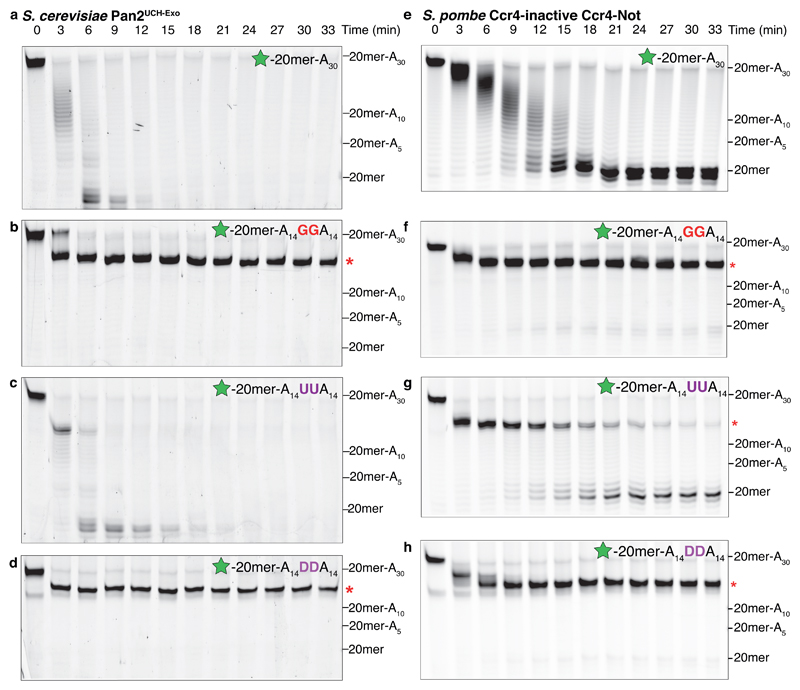
Figure 6. Nucleotide base stacking is required for Pan2 and Caf1 deadenylase activity.
Denaturing RNA gels showing deadenylation by (a-d) S. cerevisiae Pan2UCH-Exo or (e-h) S. pombe Ccr4-inactive Ccr4–Not on 5′ 6-FAM-labeled (green star) RNAs consisting of a 20mer non-poly(A) sequence (see Fig. 1a) followed by the indicated tail sequence. RNAs either had no additional nucleotides (a, e), two guanosines (b, f), two uracils (c, g), or two dihydrouracils (abbreviated D, panels d, h) in the middle of the poly(A) tail. Red asterisks indicate the point of inhibition. Both Pan2 and Caf1 were strongly inhibited by guanosines and dihydrouracils interrupting a poly(A) tail. These gels are representative of identical experiments performed 2 times. Uncropped gel images are shown in Supplementary Data Set 1.