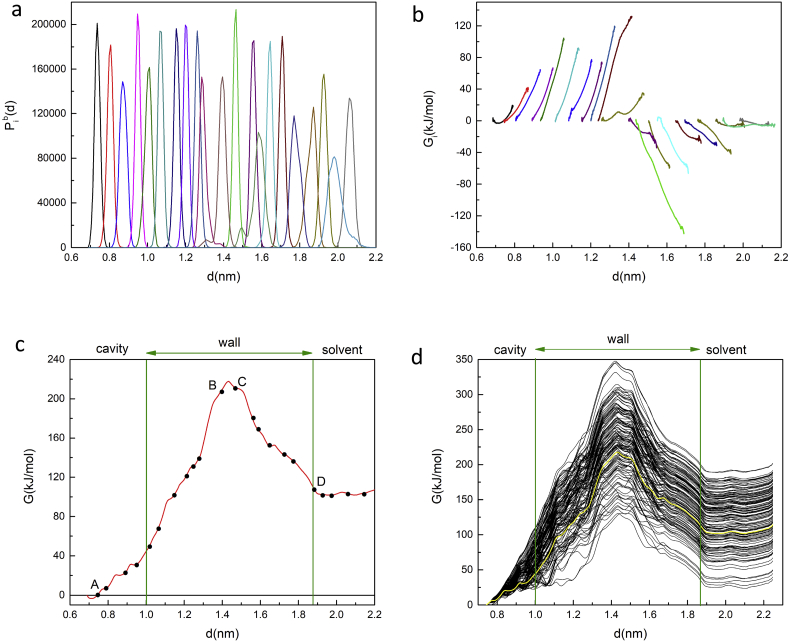
Fig. 6.
(a) The complete set of biased probability distributions of CM locations Pib(d) generated by umbrella sampling in all of the selected sampling windows along the exit trajectory of the cyclooctasulfur molecule S8 as a function of the reaction coordinate defined as the distance d of the CM of S8 relative to the location of the CM of the harmonically restrained anchor residue arginine 33 B. (b) The complete set of free energy profiles Gi(d) for all sampling windows. (c) A plot of the global free energy G(d) generated from the set of biased distributions in (a) by a simple application of WHAM. The dots show the equilibrated CM locations for all 22 sampling windows. The vertical lines mark the approximate locations of the interior and exterior surfaces of the protein wall. (d) WHAM free energy profiles generated with bootstrapping. The curve from Figure (c) is included in solid yellow. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)