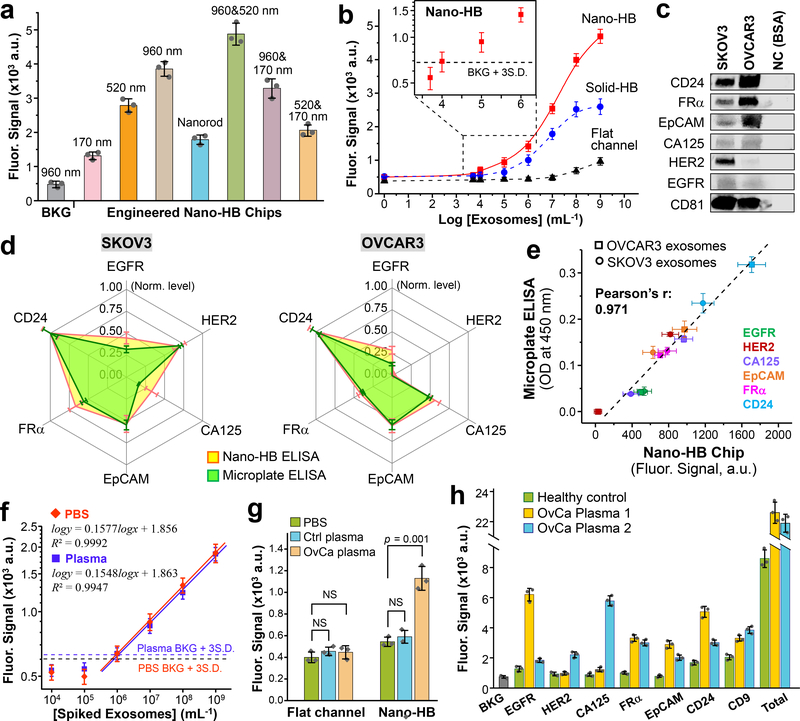
Fig. 4. Ultrasensitive detection of exosomes with the nano-HB chip.
(a) Engineering nano-HBs by the MINDS programs the sensitivity for detecting COLO-1 exosomes (105 μL−1). Statistic comparison between the 960 and 960/520 nm chips yielded p = 0.005. (b) Calibration curves for quantifying total exosomes by the flat-channel, solid-HB, and nano-HB chips. Serial 10× diluted COLO-1 exosome standards were used with a mixture of anti-CD9, CD63, and EpCAM mAbs for detection. (c) Western blot analysis of the protein markers in SKOV3 and OVCAR3 EVs with 10 μg BSA as negative control (NC). (d) Comparing the nano-HB chip and a standard microplate kit for ELISA detection of six proteins in SKOV3 and OVCAR3 exosomes. 20 μL, 105 μL−1 purified EVs were used for nano-HB assay, and 100 μL, 106 μL−1 EVs for microplate ELISA. All analyses were normalized against CD24 that was found most abundant. (e) The measurements of six targets in SKOV3 and OVCAR3 exosomes by the nano-HB chip and microplate-based ELISA correlated well. (f) Calibration curves for detecting the FRα+ subtype in UC-purified SKOV3 EVs spiked in PBS and a 10× diluted healthy plasma without detectable FRα. Total exosomes were captured with anti-CD81 mAb and the FRα+ subtype was detected with anti-FRα mAb. (g) High sensitivity of the nano-HB chip enables detection of FRα+ exosomes in an OvCa plasma sample, which is indiscernible to the conventional flat-channel chip. Two-tailed Student’s t-test was used at a significance level of p < 0.05. NS, not significant. (h) Protein profiling of exosomes directly in plasma from a control and two OvCa patients with nano-HB chips. Total exosomes were detected with a mix of CD9, CD63, and CD81 Abs. CD81 mAb was used for exosome capture in all cases. Error bars: one S.D. (n = 3).