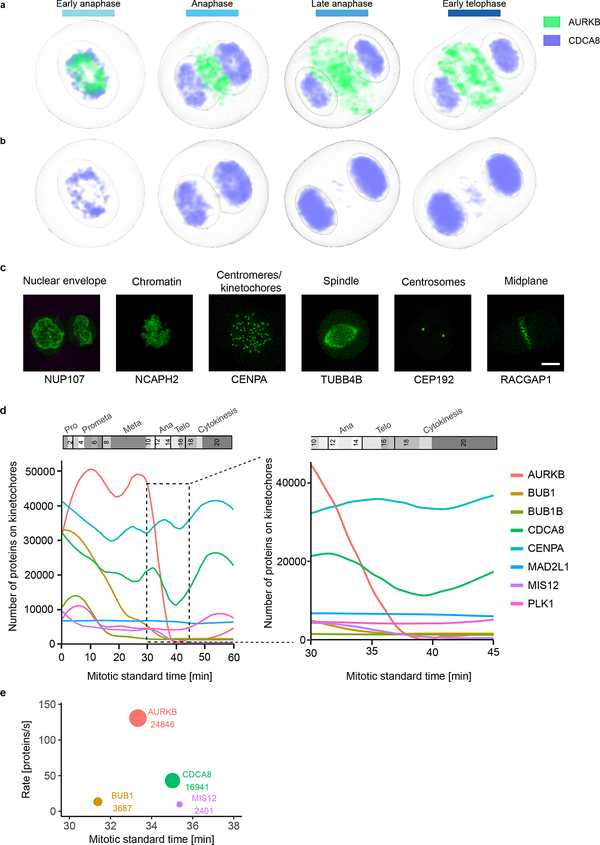
Extended Data Fig. 8 |.
Mitotic standard model and supervised classification to investigate the dynamic localization of kinetochore proteins. (a-b) Concentration maps of chromosome passenger complex proteins AURKB and CDCA8 in anaphase and early telophase. (a) AURKB concentrates in an outer ring and a central disk. Most of CDCA8 remains on chromatin and after AURKB has already relocalized, between late anaphase and early telophase, only a small CDCA8 fraction colocalizes with AURKB in the central disk. (b) Color displaying CDCA8 was adapted to make its localization in the central disk visible. (c-e) Analyzing sub-cellular (dis)assembly kinetics using a supervised approach. (c) Example of maximally Z-projected images of marker proteins for the selected subcellular compartments used for the supervised approach. Scale bar: 10 μm. (d) Kinetics of kinetochore disassembly. The predicted number of molecules localized on kinetochore/centromeres are plotted for eight proteins in the mitotic standard time (left panel) and zoomed in for anaphase (right panel). (e) Order and rate of protein removal from the kinetochore during anaphase. The annotation and circle diameter indicate the number of molecules at the estimated average time of dissociation.