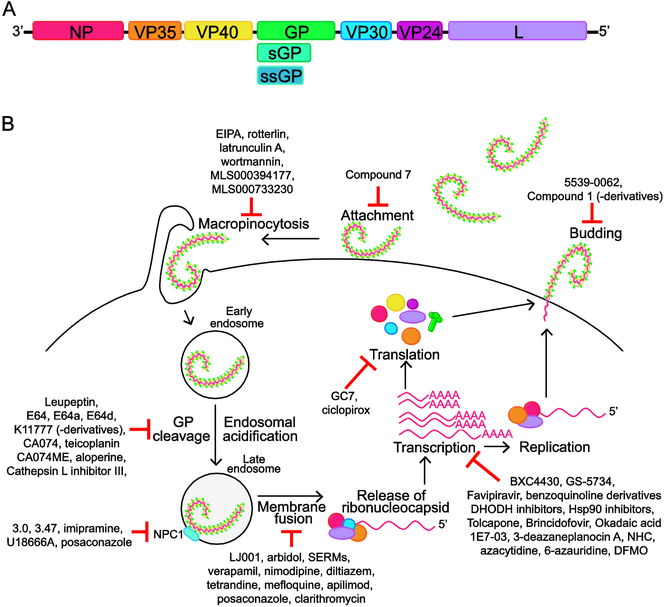
Figure 1. Filovirus genome, replication cycle and small molecules inhibitors.
A. Schematic of filovirus genome. The negative-sense RNA genome has seven transcriptional units that encode for the nucleoprotein, NP; viral protein 35, VP35; VP40; glycoprotein/soluble glycoprotein, GP/sGP (sGP is not encoded by Marburg virus); VP30; VP24; Large protein, L (viral polymerase). Note that sGP and ssGP is produced by members of the Ebolavirus genus, and predicted to be produced by LLOV. Genome schematic is not to scale. B. Schematic of the steps in the filovirus lifecycle. GP mediates attachment of the filovirus to the surface of the cell. The virus is then taken up by macropinocytosis. Following acidification of the endosome, cathepsins B and L cleave GP, a requirement for its interaction with the host protein NPC1 that facilitates fusion of viral and endosomal membranes. Endosomal calcium channels, known as two-pore channels (TPCs), play a role in trafficking the virus particle to the site of membrane fusion. Following fusion, the ribonucleocapsid is released into the cytoplasm where 5’-capped, 3’polyadenylated mRNAs are transcribed for each viral gene and a copy of the full-length genomic RNA is produced, which acts as a template for synthesis of new negative-sense viral genomes. Transcription requires NP, VP35, VP30 and L, while replication does not need VP30. Viral proteins are translated from the viral mRNAs and new viral particles are formed at the cell surface. VP40 drives viral budding and is assisted by GP. Viral ribonucleoproteins containing genomic RNA, NP, VP35, VP30 and VP24 are incorporated into the budding particles. The steps in the filovirus lifecycle are potential targets for therapeutic intervention; small molecules that target these processes are noted on the schematic. Greater detail on the filovirus lifecycle and these small molecules can be found in the review.