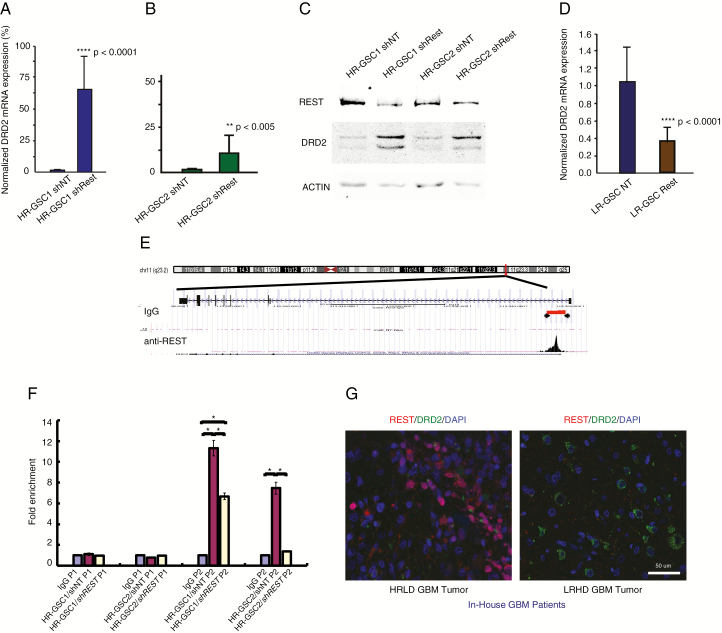
Fig. 1.
REST suppresses DRD2 gene expression in HR-GSCs. (A, B) Loss-of-function and rescue experiments. Knockdown of REST using shREST lentiviruses, compared with lentiviruses carrying the shNT control, increased DRD2 transcript levels by 65-fold in HR-GSC1 cells (A) and by 10-fold in HR-GSC2 cells (B). (C) Protein extracts from cells generated in (A) and (B) were subjected to western blot analysis. (D) Overexpression of REST in LR-GSC causes decreased DRD2 transcript levels. (E) REST binds to the DRD2 gene chromatin, as determined by unbiased genome-wide ChIP-seq assays. REST binds to a genomic region spanning 5000–5200 bp upstream of the DRD2 transcription start site (arrowheads). IgG control did not show such binding. (F) Validation of REST binding to the DRD2 gene chromatin by quantitative ChIP. REST loss-of-function HR-GSC1 and HR-GSC2 cells and their corresponding control cells as described in (A) above were processed for ChIP. REST binding to the DRD2 chromatin is shown as fold enrichment. Primer sets corresponding to the control non-binding site 4400–4600 (P1) and another set corresponding to the REST binding site 5000–5200 (P2) on the DRD2 chromatin region are shown; *P < 0.05. (G) REST-DRD2 inverse relationship exists at the protein level in our in-house GBM patient tumors. One HR- and one LR-GBM tumor specimen were analyzed by double immunofluorescence using anti-REST and anti-DRD2 antibodies. Results suggest distinct HRLD and LRHD subtypes.