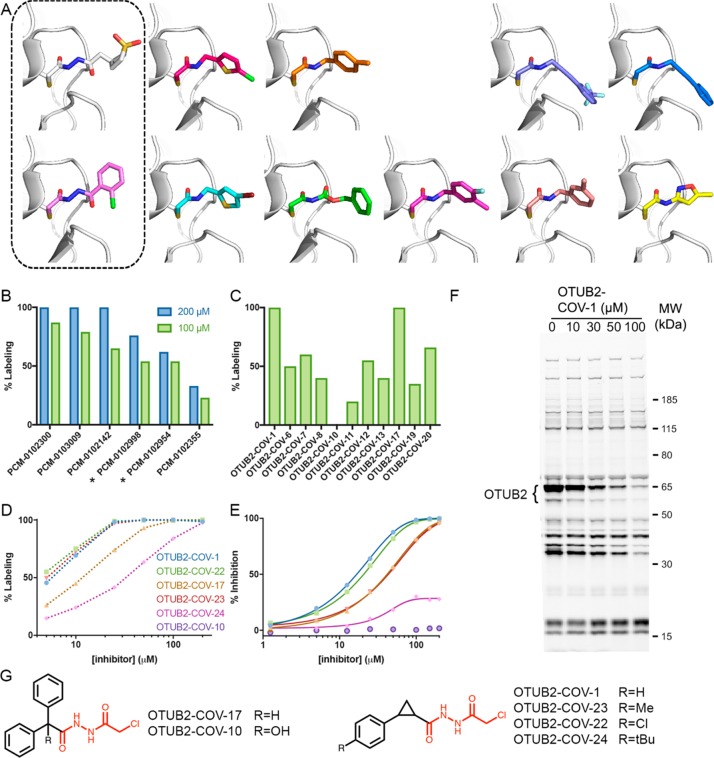
Figure 4.
Discovery of a selective OTUB2 inhibitor by fragment growing. (A) Cocrystal structures of OTUB2 in complex with (from top left) PCM-0102998, PCM-0103080, PCM-0102660, PCM-0103011, PCM-0103007, PCM-0102954, PCM-0103050, PCM-0102153, PCM-0102305, PCM-0102821, and PCM-0102500 (see Figure S13 in the Supporting Information). Structures with compounds containing the chloroacethydrazide motif are boxed. (B) Percent covalent labeling of OTUB2 with compounds containing the identified motif, PCM-0102300, PCM-0103009, PCM-0102142, PCM-0102998, PCM-0102954, and PCM-0102355 (Figure S13 in the Supporting Information), at 200 μM (blue) and 100 μM (green). Compounds boxed in (A) are marked with asterisks. (C) Percent covalent labeling of OTUB2 with selected next-generation compounds at 100 μM (see Figure S16 in the Supporting Information for all analogues and Table S4 in the Supporting Information for percent labeling). (D) Dose–response measurement of percent labeling by next-generation OTUB2 binders. All labeling in (B)–(D) was measured via LC/MS after 24 h incubation at 4 °C. (E) Inhibition of OTUB2 in an enzymatic assay (2.5 h preincubation in the presence of 2 mM free cysteine). (F) Gel-based ABPP selectivity assessment using a fluorescent activity-based DUB probe73 in HEK293 cells overexpressing OTUB2 (see Figure S21 in the Supporting Information for the full gel and additional controls). Cells were incubated with DMSO or increasing concentrations of OTUB2-COV-1 and then lysed, and the DUBs were fluorescently labeled with an alkyne ABPP probe: the higher the level of labeling by an inhibitor, the lower the level of labeling by the ABPP probe. The only bands that are reduced by OTUB2-COV-1 up to 30 μM correspond to OTUB2-GFP (at 65 kDa) and a degradation product (∼55 kDa). This demonstrates both OUTB2’s cellular engagement with OTUB2-COV-1 in cells and its high selectivity over other DUBs (see Figure S20 in the Supporting Information for the corresponding experiment in lysates). (G) Chemical structures of selected next-generation OTUB2 binders. The chloroacethydrazide motif is highlighted in red.