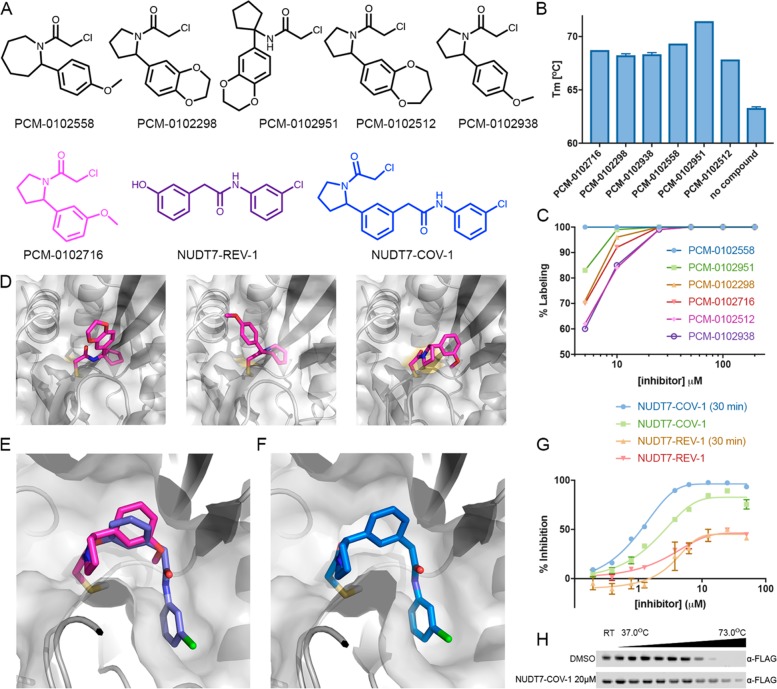
Figure 5.
Discovery of a potent NUDT7 inhibitor by fragment merging. (A) Chemical structures of similar hit compounds that exhibited 68% (PCM-0102716), 88% (PCM-0102512), and 100% (PCM-0102558, PCM-0102298, PCM-0102951, and PCM-0102938) labeling of NUDT7 in the primary screen. Compound NUDT7-REV-1 is a noncovalent fragment (purple) that was identified as a NUDT7 binder in a crystallography soaking screen (see (E)). NUDT7-COV-1 (blue) is a merged compound based on PCM-0102716 (magenta) and NUDT7-REV-1. (B) The six hits identified in the primary screen stabilize NUDT7 by 4.5–8.1 °C in a Tm shift assay. (C) Labeling percentage of compounds PCM-0102558, PCM-0102951, PCM-0102298, PCM-0102716, PCM-0102512, and PCM-0102938 at 5–200 μM, 24 h, 4 °C. (D) Cocrystal structures of NUDT7 with compounds PCM-0102951, PCM-0102558, and PCM-0102716. (E) Overlay of the crystal structures of NUDT7 with compound PCM-0102716 (pink) and with the noncovalent fragment NUDT7-REV-1 (purple). (F) The cocrystal structure of NUDT7 with the merged compound NUDT7-COV-1 adopts the exact same binding mode as the two separate fragments. (G) Enzymatic inhibition of NUDT7 by NUDT7-COV-1 and NUDT7-REV-1. The data shown include results with and without a 30 min protein preincubation in the presence of the compounds. (H) Intracellular target engagement is demonstrated by thermal stabilization of FLAG-NUDT7 by NUDT7-COV-1 in intact HEK293 cells. After transfection, cells were treated with 20 μM NUDT7-COV-1 or DMSO for 30 min before being heated to the indicated temperatures.