Figure 1.
The Generation and Characterization of HNF4α Genome-Edited Cell Lines
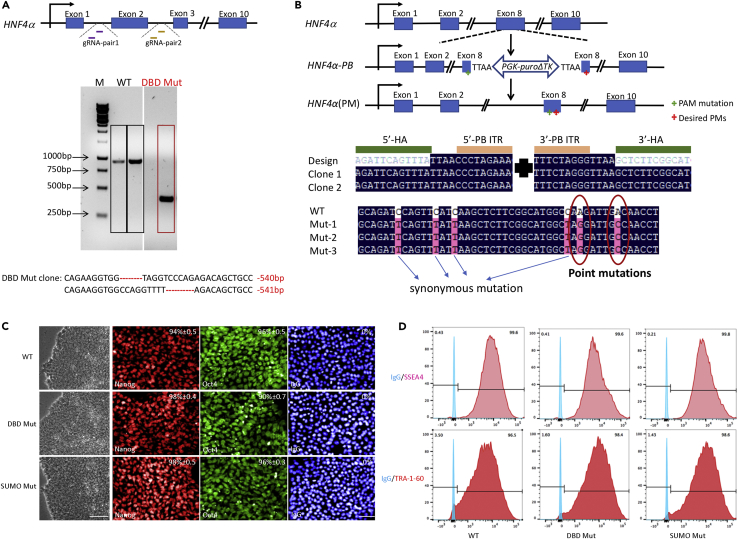
(A) Two guide RNA pairs targeting introns 1 and 2 were used to delete exon 2 in HNF4α (top panel). A homozygous deletion clone was identified by amplifying the targeted region (middle panel). Sequencing confirmed the deletion mutant (DBD Mut) clone had a 540/541-bp deletion in each allele (bottom panel). See Tables S5 and S6 for further details.
(B) A piggyBac-based targeting vector was used in combination with Cas9 nickases to introduce desired point mutations into HNF4α (top panel). The targeted clones incorporated the selection cassette (middle panel). This selection cassette is contained within the piggyBac transposon and consists of a positive-negative selection marker (puro-tk) expressed from a constitutively active promoter (PGK). Post excision of the transposon, the locus was modified seamlessly (bottom panel). PAM, protospacer-adjacent motif; HA, homology arm; PB, piggyBac, 5′-PB ITR and 3′-PB ITR are 5′ and 3′ piggyBac inverted terminal repeats flanked by the TTAA direct repeats. See Tables S5 and S6 for further details.
(C) Representative images of cellular morphology, immunofluorescences of NANOG and OCT4. One wild-type (WT) clone, one DBD Mut clone, and one point-mutated (SUMO Mut) clone was selected for characterization. IgG was used as a negative control. The percentage was calculated using four random fields of view. Scale bar, 100 μm for phase contrast and 50 μm for immunostaining images.
(D) Flow cytometry of SSEA4- and TRA-1-60-expressing cells in the WT, DBD Mut, and SUMO Mut clones. IgG was used as a negative control. N = 3 independent experiments.