Figure 3.
Frameshifting Variants in HNRNPR Truncate a Predicted PrLD in hnRNPR Protein
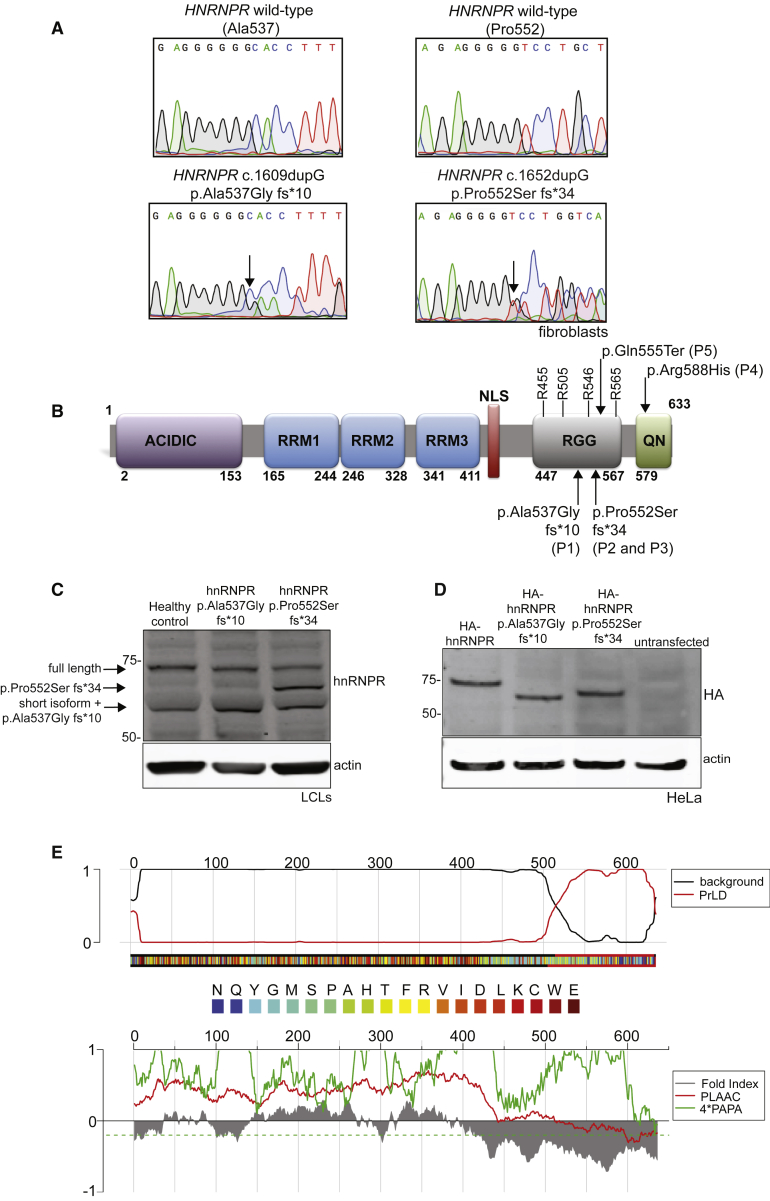
(A) Chromatograms of DNA sequencing of fibroblasts derived from healthy controls (top panels) and from P1 (p.Ala537Gly fs∗10) and P2 (p.Pro552Ser fs∗34) (bottom panels). Arrows in the bottom panels indicate the frameshift.
(B) Protein domains of hnRNPR. The acidic domain (in purple), RNA-binding domains (RRM1, RRM2, and RRM3; in blue), the nuclear localization signal (NLS; in red), the arginine/glycine domain (RRG; in gray), and the glutamine/asparagine domain (QN; in green) are illustrated. Arginine residues reported to be methylated by PRMT1 in the RGG domain are indicated, as are the positions of the variants in individuals P1–P5.
(C) An immunoblot of lymphoblast cell lines derived from a healthy control, P1, and P2 by using antibodies against hnRNPR. The two known isoforms (full length and short form) of hnRNPR are indicated by the arrows, as are the truncated proteins (hnRNPR p.Ala537Gly fs∗10 runs at the same height as the hnRNPR short form).
(D) An immunoblot of HeLa cells transfected with plasmids that coded for full-length HA-hnRNPR, HA-hnRNPR p.Ala537Gly fs∗10, and HA-hnRNPR p.Pro552Ser fs∗34 done by using antibodies against the HA tag.
(E) FoldIndex predicts an intrinsically unfolded region in the C terminus of hnRNPR (red curve reaching 1 on the top panel, gray curve below zero on bottom panel). The algorithms of Alberti et al.62 (red curve below zero on the bottom panel) and Toombs et al.63 (green curve below zero) also predict a PrLD at the C terminus of hnRNPR.