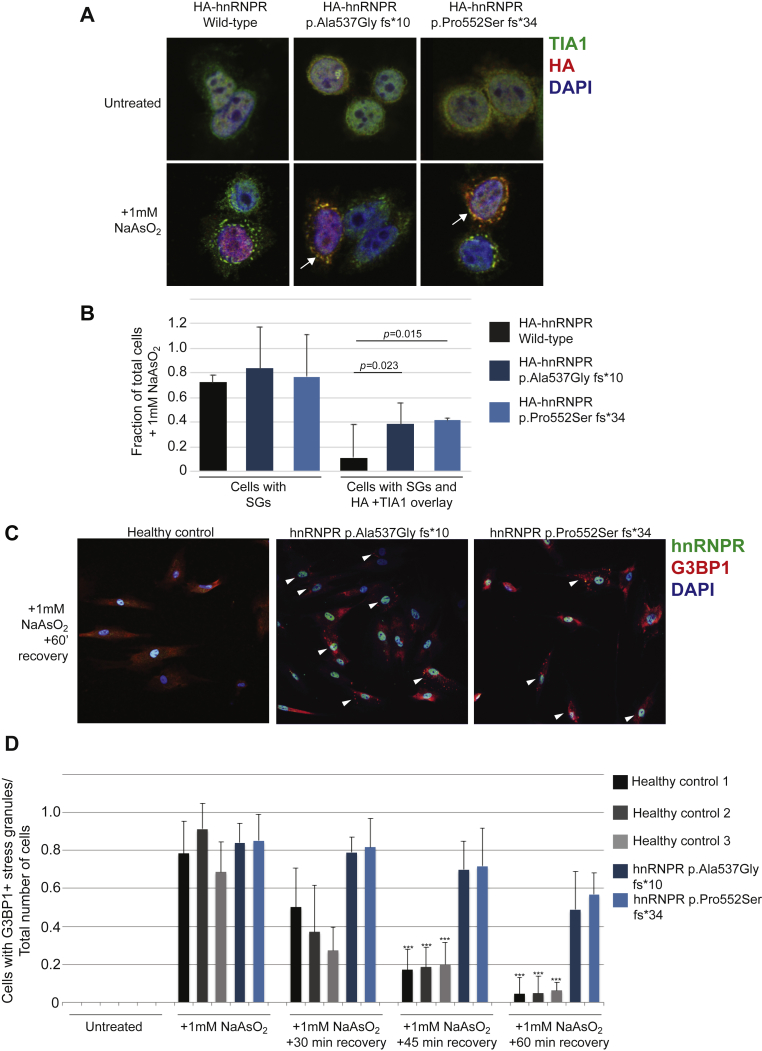
Figure 7.
Truncated hnRNPR Proteins Affect Stress-Granule Dynamics
(A) A confocal microscopy analysis of HeLa cells that were transfected with plasmids coding for HA-tagged hnNRPR full-length and truncated proteins. The antibodies that were used for staining are against the stress-granule marker TIA1 (green) and the HA tag (red). DAPI (blue) is staining the nucleus. Stress granules form upon exposure to 1 mM NaAsO2, and more cells are revealed with an overlay between TIA1 and HA-tagged truncated proteins (in yellow, indicated by arrows).
(B) A quantification of the cells in (A) (N = 3 independent transfections) that formed stress granules, as well as the cells that formed an overlay between TIA1 and HA-tagged hnRNPR proteins.
(C) A confocal microscopy analysis of fibroblasts derived from a healthy control or individuals carrying variants in HNRNPR; the fibroblasts were exposed to 1 mM NaAsO2 for 60 min and then allowed to recover for 60 min. The antibodies used for staining are against hnRNPR (green) and the stress-granule marker G3BP1 (red). DAPI (blue) is staining the nucleus. Arrowheads indicate cells revealing stress granules.
(D) A quantification of fibroblasts that formed stress granules in untreated cells, cells exposed to 1 mM NaAsO2 for 60 min and immediately fixed, and a recovery time course (N = between 3–5 independent experiments). Three unrelated cell lines from healthy controls were used (black and gray bars) for a comparison to cells carrying HNRNPR variants (blue bars). A Student’s t test was used for statistical analysis, ∗∗∗p < 0.001.
All error bars were derived using the standard deviation.