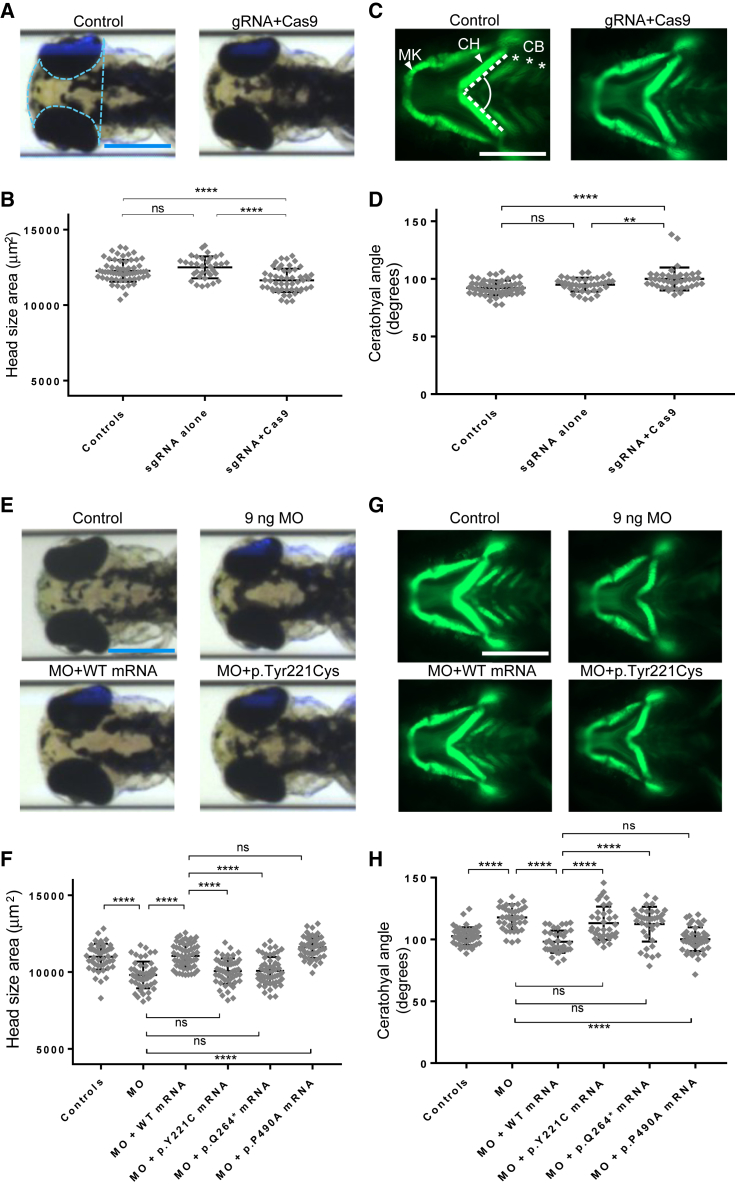
Figure 3.
Zebrafish Models of dync1i2a Disruption Display Neuroanatomical and Craniofacial Patterning Defects, and GenBank: NM_001271789.1: c.662A>G (p.Tyr221Cys) Is a Loss-of-Function Variant
(A) Representative dorsal bright field images of control and F0 mutant larvae.
(B) Quantification of head size area in F0 mosaic mutants
(C) Representative ventral images of the GFP signal in the anterior region of -1.4col1a1:egfp transgenic reporters for control and F0 mutant larvae. Abbreviations are as follows: MK = Meckel’s cartilage; CH = ceratohyal cartilage; and CB = ceratobranchial arches.
(D) Quantification of the CH angle in F0 mutants
(E) Representative dorsal bright field images of control, e2i2 morphant (MO), WT, or mutant mRNA rescue larvae.
(F) Quantification of complementation assays that used head size as a phenotype readout; p.P490A is a negative control variant (dbSNP: rs767705533, 2 homozygotes in ∼120,000 individuals in gnomAD)
(G) Representative ventral images of the GFP signal in the anterior region of -1.4col1a1:egfp transgenic reporters for control, e2i2 morphant, WT, or mutant mRNA rescue larvae.
(H) Quantification of complementation assays that used the CH angle as a phenotype readout; data are concordant with the scoring of head size. All phenotyping was performed at 3 days post-fertilization.
The pale blue dotted line in panel A indicates the area measured to obtain the data in panels B and F. The angle indicated between the dashed white lines in panel C indicates the measurement used to obtain data for panels D and H. Scale bars in panels A, C, E, and G represent 300 μm; image sizing is consistent across panels. ∗∗∗∗p < 0.0001; ∗∗p < 0.01; and ns = not significant; n = 40–67/condition, and three biological replicates showed similar results for data shown in panels B, D, F, and H. Error bars represent standard deviation of the mean. Note that the amino acid codons Y221C, Q264∗, and P490A of the most abundant isoform (GenBank: NM_001271789.1) used in the zebrafish experiments correspond to Y247C, Q290∗, and P516A of the longest isoform (GenBank: NM_001378.2) of DYNC1I2.