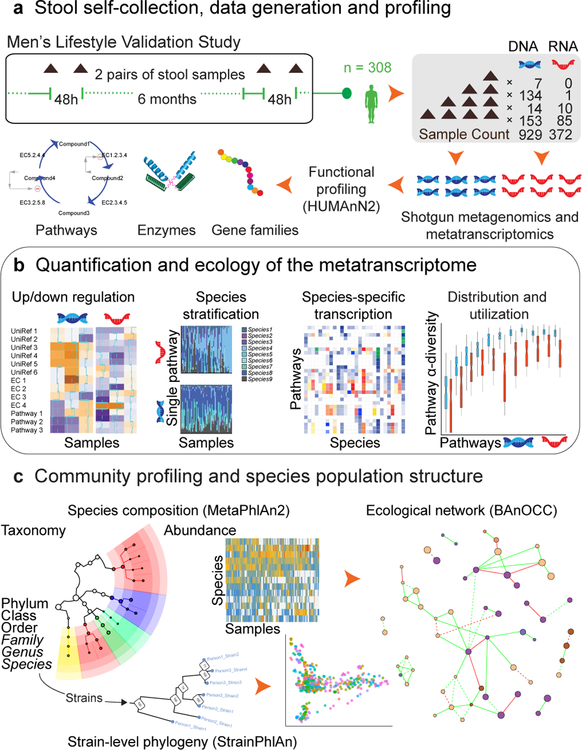
Figure 1. Metatranscriptomic and metagenomic taxonomic and functional profile of a prospective human cohort.
A) 308 participants from the Men’s Lifestyle Validation Study (MLVS), embedded within the Health Professionals Follow-up Study (HPFS) prospective cohort13, provided a target of four stool samples each. These were self-collected in two pairs, six months apart, with each pair spanning 2–3 days. This yielded 929 total metagenomes and 372 metatranscriptomes, sequenced using previously published protocols9 and functionally profiled using HUMAnN215. B) To estimate gene family, enzyme class, and pathway relative transcription, RNA abundances were normalized to corresponding DNA abundances. We then evaluated “core” (prevalently transcribed) and variable transcriptional elements, in addition to the ecological and phylogenetic diversity of metatranscription and carriage of functional elements among species. C) Taxonomic profiles were determined using MetaPhlAn214 from both DNA and RNA data (for RNA viruses). These were also used for ecological interaction network reconstruction25 with BAnOCC (Schwager in review) (http://huttenhower.sph.harvard.edu/banocc) and for strain tracking with StrainPhlAn28.