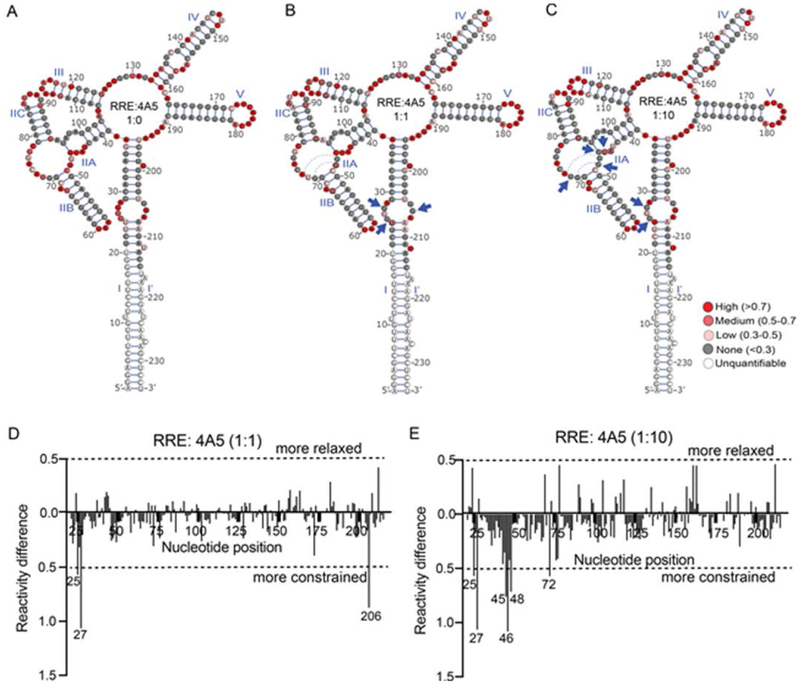
Figure 4.
SHAPE-MaP reactivity profiles of 234 nt RRE RNA in the presence of peptide 4A5. Upper panel:Reactivity values of the RRE superimposed on the 5 stem-loop structure of NL43 RRE (A) in the absence of 4A5, (B) in the presence of equal molar concentration of 4A5, and (C) in the presence of 10-fold molar excess of 4A5. The solid blue triangles point to nucleotides whose reactivity changes significantly in the presence of 4A5. Lower panel: Reactivity difference profile generated for (D) equimolar RRE:peptide ratio and (E) 1:10 RRE:peptide molar ratio by subtracting reactivity values of the RRE in the absence of the 4A5 from those in the presence of 4A5. Here +/− 0.5 difference is used as cut-off value to identify nucleotides that are more relaxed or more constrained in the presence of 4A5. Dotted blue arcs represent the non-canonical bonds shown to form upon Rev binding.