Extended Data Figure 9. Genome-wide associations between H3K4me3Q5ser and TFIID.
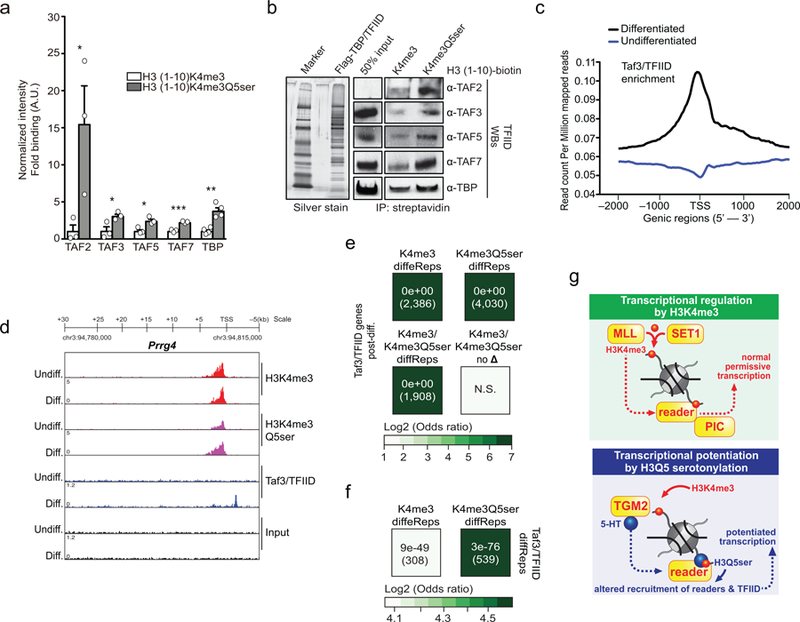
a, Western blot quantifications related to Fig. 4a (Student’s t-tests, two-sided) of modified H3 (1–10) peptide IPs from HeLa nuclear extracts (TAF2 – n=3/peptide; t4=2.724, *p=0.05, TAF3 - n=3/peptide; t4=2.920, *p=0.04, TAF5 - n=3/peptide; t4=3.685, *p=0.02, TAF7 - n=3/peptide; t4=8.885, ***p=0.0009, TBP – n=4/peptide; t6=5.383, **p=0.0017). b, Modified H3 (1–10) peptide IPs against purified TFIID from Flag tagged TBP expressing soluble HeLa nuclear extracts (left, silver stain of the purified complex), followed by WBs (right). Inputs are provided (n=1/protein examined). c, Gene plot (ngs.plot) of Taf3/TFIID enrichment (Log2 fold change vs. input) comparing signals pre- vs. post-differentiation (n=3 independent biological replicates/differentiation states). d, IGV genome browser tracks of the Prrg4 locus for H3K4me3, H3K4me3Q5ser and Taf3/TFIID (vs. DNA input) in RN46A-B14 cells pre- and post differentiation (example locus was chosen based upon MACSv2.1.1- and diffReps-based statistical comparisons). Odds ratio analysis (using Fisher’s exact tests) of overlapping genes displaying differential H3K4me3 or H3K4me3Q5ser enrichment (or those genes containing differential sites for both or neither of the two marks, FDR<0.05, FC>1.2) vs. e, Taf3 peaks (n=3 independent biological replicates/differentiation state, MACSv2.1.1, FDR<0.05, FC>1.2 cutoffs applied after adjusting for multiple comparisons; normalized to respective DNA inputs) or f, differential Taf3/TFIID enrichment (n=3 independent biological replicates/differentiation state, diffReps, FDR<0.05, FC>1.2 cutoffs applied after adjusting for multiple comparisons) post-differentiation. Insert numbers indicate respective p values for associations, followed by the number of protein-coding genes overlapping per significant category. g, Model of the impact of Q5ser on K4me3-mediated gene expression in vivo – our data suggest that the presence of combinatorial H3K4me3Q5ser alters interactions with certain K4me3 “reader” proteins, such as the TFIID complex, to potentiate/stabilize permissive gene expression in mammalian cells.
