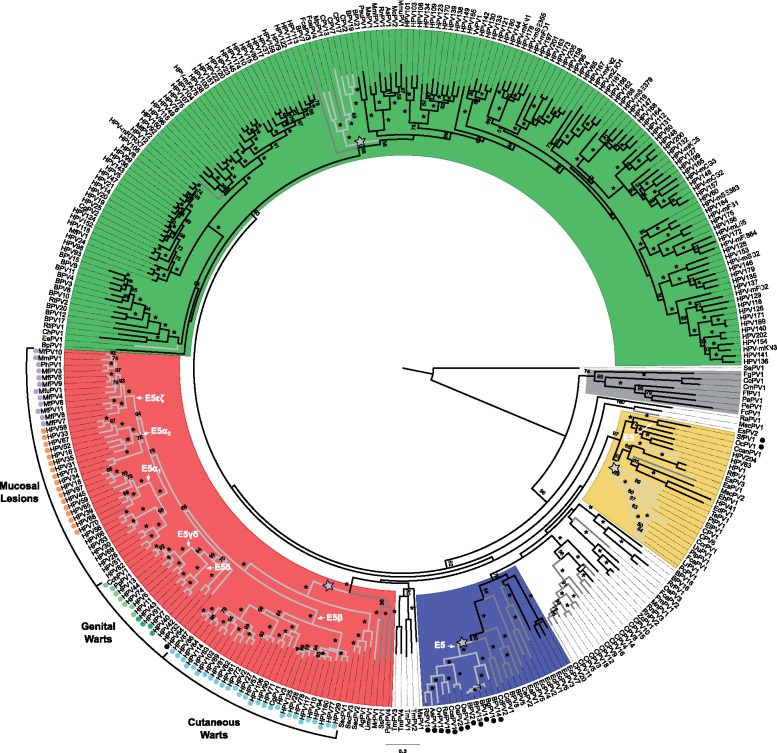
Fig. 1.
—Best-known maximum likelihood phylogenetic tree of the concatenated E1E2L2L1 amino acid sequences of 343 PVs and identification of clades containing an intergenic E2–L2 region. Clade color codes highlight the four PV crown groups: red, Alpha-OmikronPVs; green, Beta-XiPVs; yellow, Lambda-MuPVs; blue, Delta-ZetaPVs; gray, a yet unclassified crown group consisting of PVs infecting birds and turtles; and white, PVs without well-supported phylogenetic relationships. Outer labels, Mucosal Lesions, Genital Warts, and Cutaneous Warts, indicate the most common tropism for the AlphaPVs. Values on branches correspond to ML bootstrap support values. Asterisks indicate a maximal support of 100, and values under 50 are not shown. Branches in light-gray correspond to PV genomes containing an inter-E2–L2 region with a minimum size of 250 nt; branches in dark-gray correspond to PV genomes with an inter-E2–L2 region with a minimum size of 125 nt. The basal nodes of the four clades containing a relatively long intergenic region between the E2 and the L2 ORFs are labeled with a star. The basal node of the lineages containing an E5 coding sequence is indicated with an arrow, and the corresponding terminal taxa are labeled with a color-coded dot indicating the E5 type. Purple dots indicate: E5, orange dots: E5α, light green dots: E5γδ, dark green dots: E5δ, blue dots: E5β, and black dots are lineages containing unclassified E5 types.