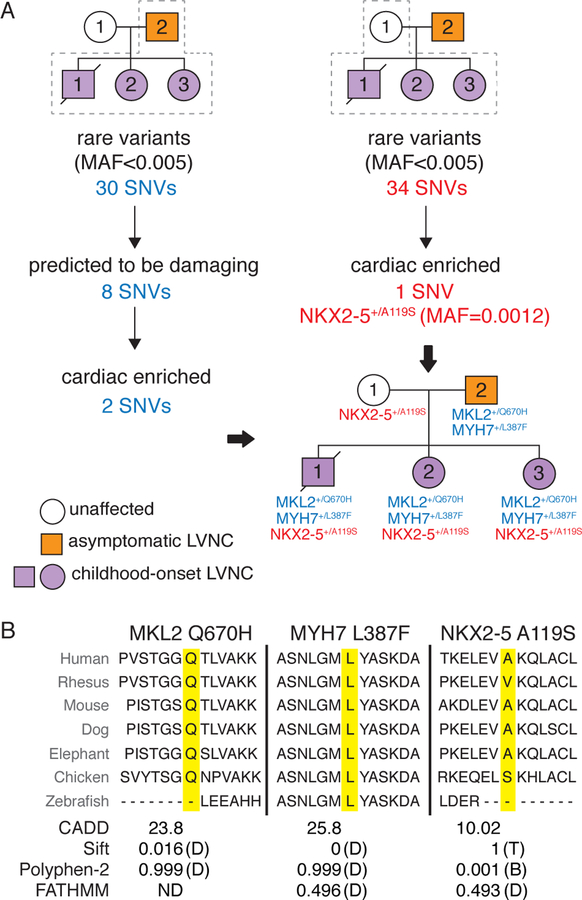
Fig. 2. Genotypic analysis of LVNC family by exome sequencing.
(A) Workflow for analysis of sequence variants inherited from the father is shown in blue on the left and from the unaffected mother in red on the right. Inheritance pattern for variants of interest is shown in pedigree on lower right. (B) Conservation of each amino acid residue across species is shown. The variant of interest is highlighted in yellow. Predicted effects of variants on protein function based on Combined Annotation Dependent Depletion (CADD), Sift, Polyphen-2 and FATHMM programs. Damaging (D), benign (B), tolerated (T) and not determined (ND). Single nucleotide variant (SNV); Minor allele frequency (MAF).