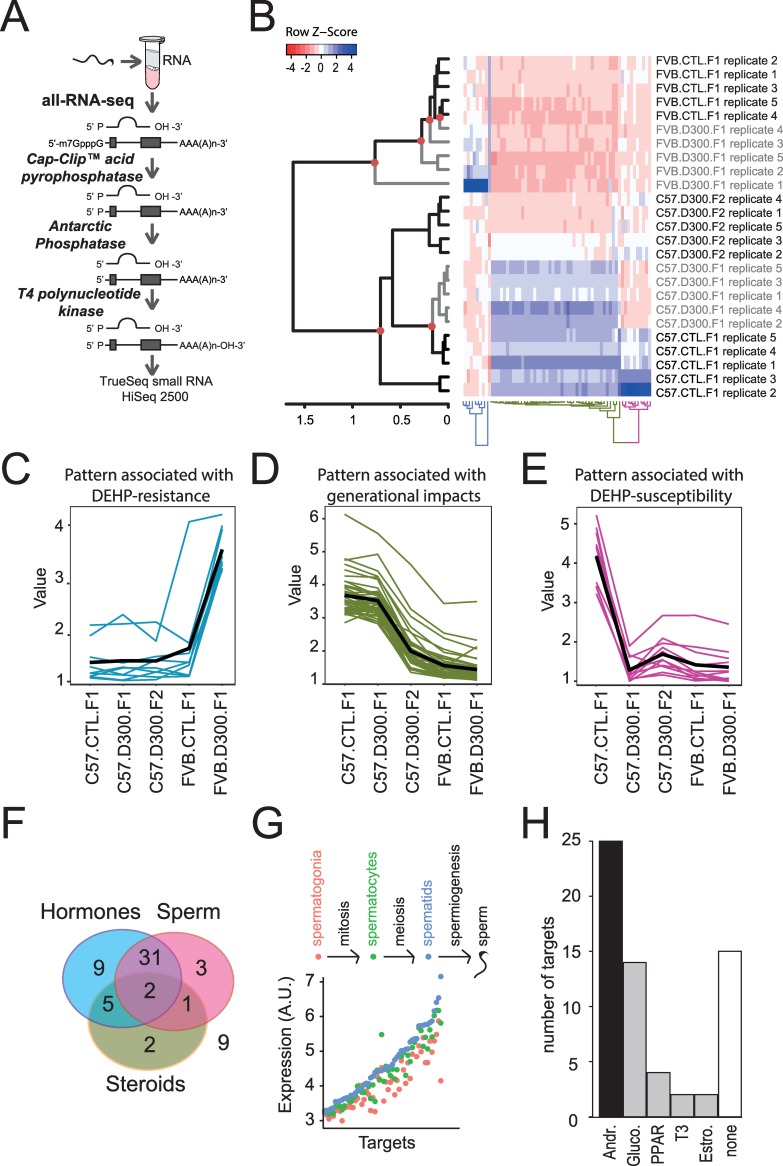
Fig 2. Results obtained in the genome*environment*generation transcriptomic interactions study.
(A) All-RNA-seq analysis in sperm. Sperm sampled from the cauda epididymis was analyzed in quintuplicates using the all-RNA-seq protocol, which allows measurements of all RNA types independently of their end variations. (B) Hierarchical clustering analysis combined with a heat-map illustration of all-RNA-seq data. Red circles: significant nodes in the dendrogram. In the heat-map, high levels of expression are in blue and low levels in red. (C) The expression pattern associated with the DEHP-resistance indicates induction of expressions in FVB.D300.F1 compared with FVB.CTL.F1. (D) Pattern associated with generational impacts in the C57BL/6J background. Decreased expressions are observed in C57.D300.F2 compared with C57.CTL.F1 and C57.D300.F1. (E) The expression pattern associated with DEHP-susceptibility displayed decreased expressions in C57.D300.F1 compared with C57.CTL.F1. (F) Triple-Venn diagram segregating the 62 RNAs. The 62 RNAs were segregated in a Triple-Venn diagram as hormonally-regulated (light blue circle: N = 47; 76%), as previously identified in sperm (pink circle: N = 37; 60%), or involved in the transport of cholesterol or the metabolism of steroids (yellow circle: N = 10; 16%). The majority of RNAs are hormonally-regulated and expressed in sperm (N = 33; 53%); the minority of RNAs are out of the diagram (N = 9; 15%). (G) Expression levels of RNA across germ cells subtypes: spermatogonia in red, spermatocytes in green, and spermatids in blue. RNAs were significantly enriched for increased expression in spermatogenesis compared with all RNAs in the Chalmel dataset (χ2(1) = 53; p< 3.3*10−13). (H) Number of RNA regulated by androgen (Andr.), glucocorticoids (Gluco.), hormone-like PPARs, triiodothyronine (T3), estrogen (Estro.) or not regulated by hormones (none). 76% of the targets present a reported hormonal regulation (47/62), among these 53% by androgen (25/47).