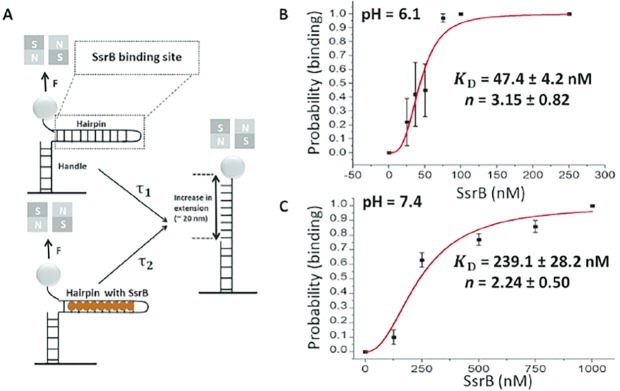
Figure 8. Quantification of SsrB-DNA binding affinity.
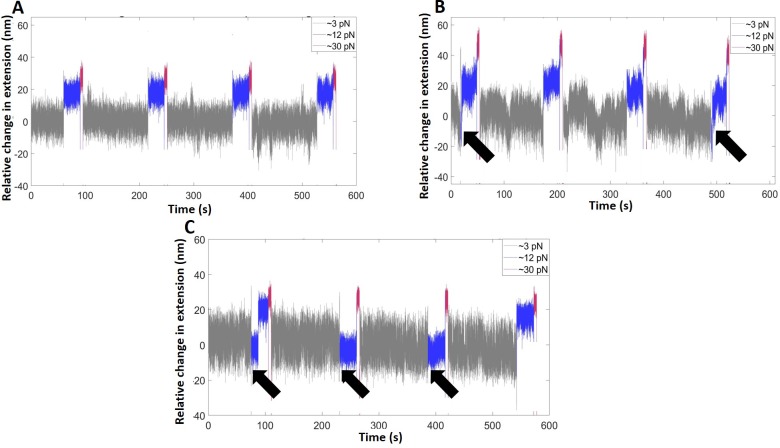
(A) Schematic of the principle of the measurement (see Gulvady et al., 2018 for more details). A force is applied to the hairpin using a pair of permanent magnets shown in gray (N: north pole, S: south pole). At a force slightly greater than the critical force, Fc, the naked DNA hairpin has a short lifetime of τ1, while the hairpin bound with SsrB has a much longer lifetime of τ2. This delayed unzipping indicates SsrB binding. See Figure 8—figure supplement 1 for the binding traces. (B) The equilibrium binding probability as a function of SsrB concentration was plotted at pH 6.1 and (C) pH 7.4. These were the measured intracellular pH values of Salmonella in response to acid or neutral pH (Chakraborty et al., 2015; Chakraborty et al., 2017). The solid curves are the fitted curves to the Hill equation, the error bars represent the S.E.M. At pH 6.1 the KD was 47.4 ± 4.2 nM and the Hill coefficient (n) was 3.15 ± 0.82 nM. At pH 7.4, the KD increased to 239.1 ± 28.2 nM and the Hill coefficient (n) was still cooperative at 2.14 ± 0.5. Three to five independent tethers were analyzed for each point of the curves, and at least three independent probability values were determined at each SsrB concentration.