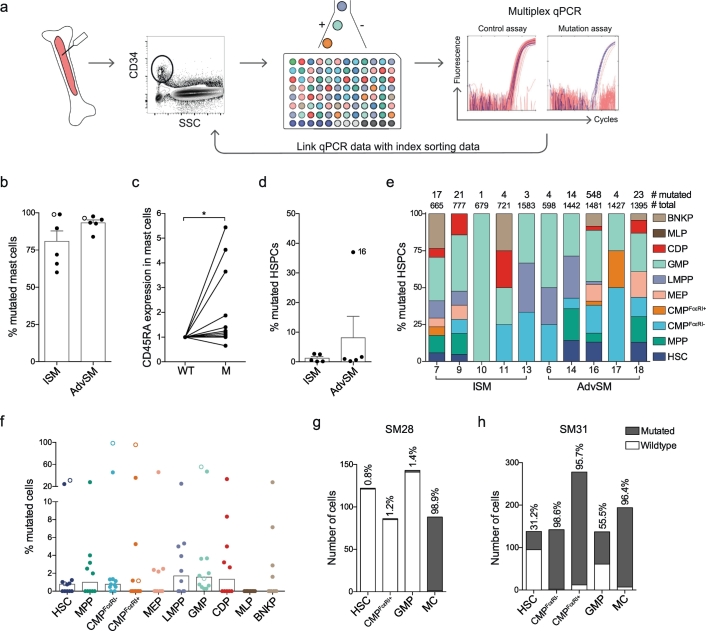
Fig. 4.
Mutation analysis of KIT D816V in single cells identifies mutated HSPCs and mast cells. (a) Method of single-cell mutation analysis using index sorting and multiplex qPCR. (b) The bars indicate the means + SEM percentage of mutated mast cells. (c) CD45RA expression of mutated and wildtype (WT) mast cells was determined and normalised to the expression in wildtype cells for each patient. Eleven patients are shown, as index data was not available from 1 of the 12 patients. The numbers of cells analysed from each patient are presented in Supplementary Table 2. (d) The bars indicate the means + SEM percentage of mutated HSPCs. (e) The cellular distribution of the mutation in 10 different HSPCs, showing the total numbers of mutated and sorted cells for each sample that passed the quality control procedure. (f) The bars indicate the median percentage of mutated cells per HSPC. (g-h) Mutation frequencies of selectively isolated HSPCs following a 2-step sorting protocol in two samples (SM28, ISM; and SM31, AdvSM). These two samples are indicated with open circles in panels b and f. The two-tailed Wilcoxon matched-pairs signed rank test was used in panel c, *P < .05.