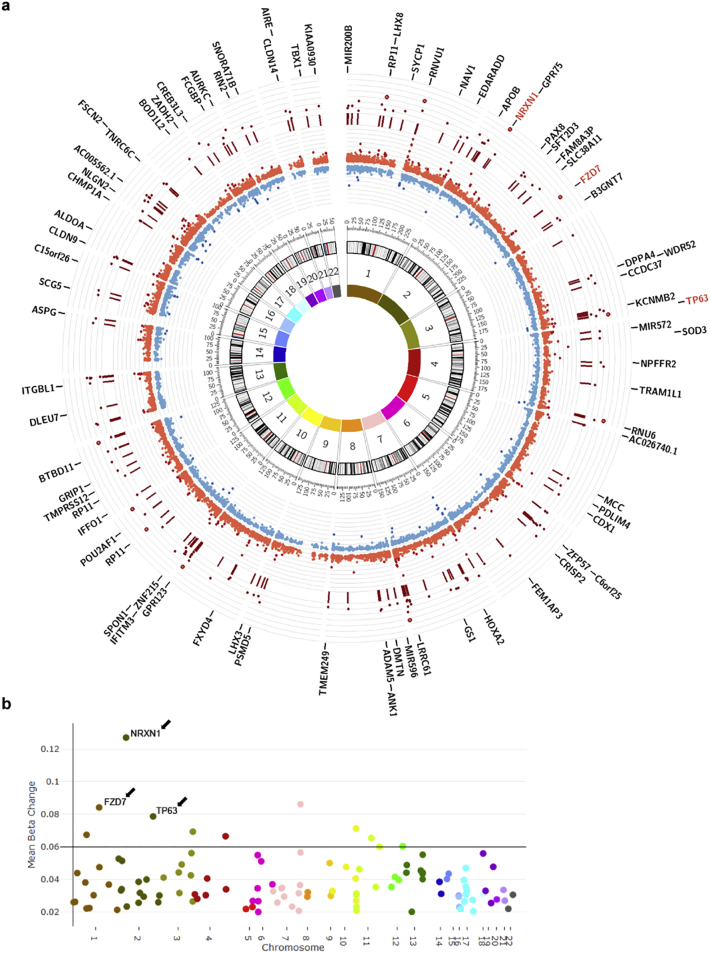
Fig. 5.
Chromosomal distribution and identification of genes with most significantly hypermethylated regions in MS patients with high BMI identifies negative regulators of cell proliferation. (a) Circos plot representing the chromosomal distribution of DMRs and CpGs within DMRs. Chromosomal numbers are indicated in the innermost circle. The inner red and blue circle depict the beta value differences for each CpG within a DMR. The more hypermethylated the CpG (bright red) the farther it is from the center of the circos plot, and the more hypomethylated the CpG (blue) the closer it is to the center of the plot. Dark red lines in the outer circle denote the genomic location of the DMRs and dark red dots indicate the mean beta value difference for each DMR. Genes overlapping DMRs are indicated in the outermost circle and genes with the greatest methylation differences are highlighted in red. (b) Manhattan plot highlighting the overall methylation difference, or mean beta change, for each DMR on each chromosome. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)