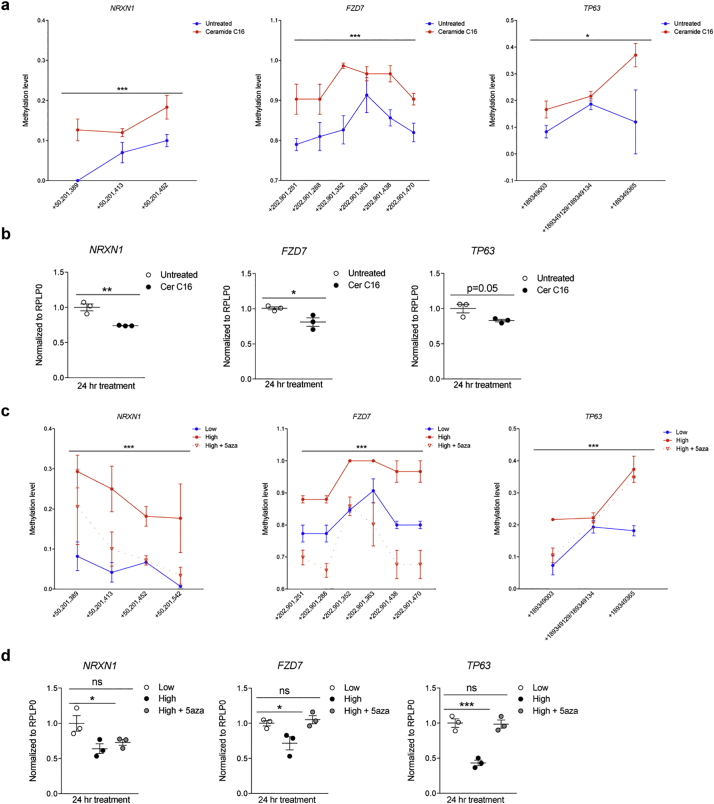
Fig. 6.
Exposure to ceramides results in increased DNA methylation and reduced transcription of negative regulators of cell proliferation. (a) Cultured human monocytes (i.e. THP1 cells) were either untreated (blue) or treated with Ceramide C16 (red) for 24 h. DNA was harvested and processed for Sequenom Mass Array EpiTyper, in order to quantify differential methylation of CpGs (Y axis) at the indicated genomic locations (X axis). The gene name is shown on top of each graph. (b) samples from the same cells described in (a) were used for RNA isolation and quantification of the corresponding gene transcripts by using real time-PCR and normalized to reference gene transcripts. (c) Cultures were treated for 24 h with concentrations of a mixture of ceramides C16, C22, and C24:1, mimicking those detected in the plasma of MS patients with normal (Low) or high BMI (High) in the absence or presence of the DNA methylation inhibitor, 5-aza-2′-deoxycytidine (5-aza). DNA was harvested and processed for Sequenom Mass Array EpiTyper, in order to quantify differential methylation of CpGs (Y axis) at the indicated genomic locations (X axis). The gene name is shown on top of each graph. Overall methylation differences for the entire region of interest for each gene were assessed using two-way ANOVA (*p < .05, ***p < .0001) (n = 3/group, n = 2 replicates/group). (d) Transcript levels of NRXN1, FZD7, and TP63 were assessed using real time-PCR and normalized to the levels of thehousekeeping gene, RPLP0. Cer C16 groups were compared to Untreated groups using Student's t-test, while the High and High +5-aza groups were compared to the Low group, using Dunnett's multiple comparisons test (*p < .05, **p < .01, ***p < .001) (n = 3 independent experiments, n = 3 biological replicates/experiment, n = 3 technical replicates/biological replicate). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)