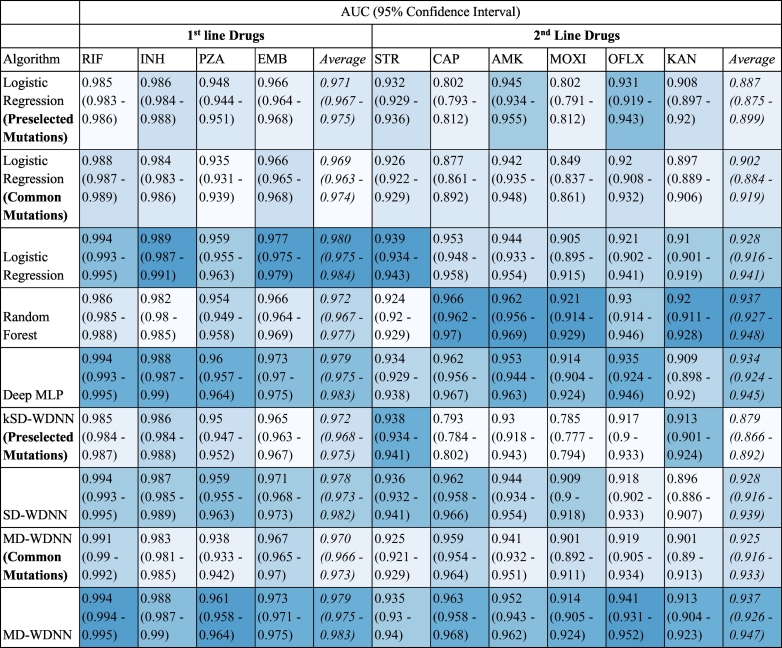
Table 1.
Tuberculosis drug resistance prediction AUROC performance of the models examined using repeated cross-validation. A table of predictive performance across all nine models during repeated cross-validation. The MD-WDNN, SD-WDNN, deep MLP, random forest, and logistic regression models were trained on the full set of predictors. The MD-WDNN (Common Mutations) and logistic regression (Common Mutations) models were trained on mutations not including the derived categories. The kSD-WDNN (Preselected mutations) and logistic regression (Preselected mutations) models were trained on preselected mutations known to be determinants of resistance for each drug. Performance is shown in average AUC and 95% confidence interval across all cross-validation folds. The cells are colored by rank of the model for each drug, colored from lightest to darkest corresponding with lowest to highest AUC value.