Abstract
Background
In our preliminary screening, expression of miR-338-5p was found to be higher in primary colorectal cancer (CRC) with metastasis. The autophagy related gene- phosphatidylinositol 3-kinase, catalytic subunit type 3 (PIK3C3) appeared to be targeted by miR-338-5p. Here, we provide solid evidence in support of PIK3C3 involved in miR-338-5p related metastasis of CRC in vitro and in vivo.
Methods
The potential clinical relevance of miR-338-5p and its target gene was analysed on benign colorectal polyps and primary CRCs by QPCR. Mouse spleen xenograft experiment was performed to examine the importance of miR-338-5p for metastasis.
Findings
PIK3C3 was one of target genes of miR-338-5p. In primary CRCs, expression of miR-338-5p is positively related to tumour staging, distant metastasis and poor patient survival. Patients with higher ratios of miR-338-5p/PIK3C3 also had significantly poor overall survival, supporting their significance in the progression of CRC. Over-expression of miR-338-5p promotes CRC metastasis to the liver and lung in vivo, in which PIK3C3 was down-regulated in the metastatic tumours. In contrast, overexpression of PIK3C3 in miR-338-5p stable cells inhibited the growth of metastatic tumours. Both migration and invasion of CRC in vitro induced by miR-338-5p are mediated by suppression of PIK3C3. Using forward and reverse approaches, autophagy was proved to involve in CRC migration and invasion induced by miR-338-5p.
Interpretation
MiR-338-5p induces migration, invasion and metastasis of CRC in part through PIK3C3-related autophagy pathway. The miR-338-5p/PIK3C3 ratio may become a prognostic biomarker for CRC patients.
Fund
NCKU Hospital, Taiwan, Ministry of Science and Technology, Taiwan.
Keywords: miR-338-5p, PIK3C3, Autophagy and colorectal cancer
Research in context.
Evidence before this study
Recent studies show that microRNA miR-338-5p suggested as a potential diagnostic biomarker for colorectal cancer (CRC). Expression of miR-338-5p was higher in CRC patients with metastasis. Phosphatidylinositol 3-kinase, catalytic subunit type 3 (PIK3C3) might be target gene of miR-338-5p.
Added value of this study
Her, we found that PIK3C3 is one of seven target genes of miR-338-5p. Expression of PIK3C3 is negatively correlated with miR-338-5p in primary CRCs. Patients with higher ratios of miR-338-5p/PIK3C3 have a significantly poor overall survival. MiR-338-5p promotes metastasis of stable cells in vivo through inhibition of PIK3C3. We also proved that cell migration and invasion activated by miR-338-5p in vitro is partially ascribed to PIK3C3 mediated autophagy. MiR-338-5p and PIK3C3 play an important role in tumour progression of colorectal cancer.
Implications of all the available evidence
MiR-338-5p/PIK3C3 ratio is a promising prognostic biomarker for CRC patients. PIK3C3 is a potential novel therapeutic target for human CRC.
Alt-text: Unlabelled Box
1. Introduction
Colorectal cancer (CRC) is the third most common human cancer worldwide and was responsible for 832,000 cancer-related deaths in 2015 [1]. The conventional paradigm of sporadic CRC arises from adenomatous polyps with progression through high-grade dysplasia to invasive CRC [2]. Tumour staging at diagnosis is the most important prognostic factor for CRC patients and is the basis for choosing an appropriate treatment regimen. As a rule, both stage I and stage II CRC patients are recommended for surgical resection, but stage III patients are treated by surgery combined with adjuvant chemotherapy [2,3]. Despite advances in neoadjuvant and adjuvant therapies, about half of CRC patients will develop recurrent disease and some of them will progress to metastatic disease. These facts indicate that conventional stage classification is not sufficient for predicting the natural course of CRC patients, nor is any biomarker a sufficiently accurate prognostic factor. Therefore, identifying an accurate prognostic marker is mandatory for CRC patients.
Recent advances suggest that microRNAs (miRNAs) warrant investigation as prognostic biomarkers for CRC patients. For example, it was reported [4,5] that miR-21 was upregulated in CRC and could be a diagnostic and prognostic serum biomarker for CRC patients. The miR-17, miR-31, and miR-126 were reported [[6], [7], [8], [9]] to be prognostic biomarkers for patients undergoing chemotherapy or anti-EGFR therapy. Using an in-house miRNA microarray, upregulated miR-338-5p was found to associate with recurrence and tumour metastasis in our pilot study [10]. MiR-338 belongs to family of brain-specific miRNA precursors [11,12] in an intronic region within apoptosis-associated tyrosine kinase (AATK) gene [13], and is upregulated in CRC [14]. The miR-338 stem loop contains miR-338-3p and miR-338-5p. In human glioma, miR-338-5p expression promotes cell invasion [15]. Increased level of serum miR-338-5p was detected in the advanced stages of patients [16], and was recently suggested as a potential diagnostic biomarker for CRC [17]. However, clinical relevance and mechanisms underlying miR-338-5p in the pathogenesis of human CRC are still unclear.
Phosphatidylinositol 3-kinase (PIK3-kinase) contains three classes of catalytic subunits: class I, class II, and class III [18]. Phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3) is important in intracellular membrane trafficking [19] and is encoded by the yeast VPS34 gene [20]. PIK3C3 could induce autophagy nucleation through complex formation by phosphorylation of 3-OH of phosphatidylinositol to phosphatidyl-inositol-3-phosphate [21]. In addition, PIK3C3 complex inhibits the epithelial-mesenchymal transition (EMT) by activating autophagy and degrading Snail and Twist in breast cancer cells, resulting in suppression of cell migration, tumour formation, and metastasis [22,23]. Autophagy has been shown to inhibit migration of cancer cells in many studies. For example, induction of autophagy repressed cervical cancer cell migration and invasion through inhibition of VEGF and MMP9 [24]. Autophagy also regulates cell migration through degradation of β1 integrin [25]. Up-regulated Beclin1 inhibits the migration and metastasis of CRC in vitro through autophagy [26]. The miR-338 may relate to autophagy. The miR-338-3p inhibits autophagy of human cervical cancer cells through PI3K/AKT/mTOR signaling pathway [27]. However, the potential significance of miR-338-5p for autophagy is unclear.
In our pilot study, transient transfection experiment of miR-338-5p suggested that PIK3C3 may be one of potential target genes [28]. To verify this hypothesis, an independent computational target prediction strategy incorporating new website was performed in cooperation with bioinformatic analysis. The objective of this study is to clarify the role, as well as the underlying mechanisms involves in PIK3C3 and autophagy, of miR-338-5p in the metastasis of CRC, and to assess the potential of miR-338-5p and PIK3C3 as a biomarker for CRC.
2. Materials and methods
2.1. Tumour tissue
A total of 29 colorectal polyps and 66 colorectal cancer specimens were collected from National Cheng Kung University Hospital (NCKUH). Specimens were collected between 2006 and 2018. This study was approved by the NCKUH Institutional Review Board (B-ER-103-031). Informed consent was signed by all patients.
2.2. Cell transfection and lentivirus infection
The cells were transiently transfected with miR-338-5p mimic (mirVana miRNA mimic) (Ambion; Invitrogen), miR-338-5p inhibitor (anti-miR-338-5p) (mirVana miRNA inhibitor) (Ambion; Invitrogen) and pCMV-Vps34 plasmid (a generous gift from WC Su, MD, China Medical University, Taiwan) using Lipofectamine 2000 (Invitrogen). p-CMV2 (Invitrogen) was chosen as the control vector.
The shRNAs targeting LacZ, GFP, ATG-5, and PIK3C3 (Vps34) were purchased from the National RNAi Core Facility (Academia Sinica, Taipei, Taiwan). The IDs of the selected clones are: TRCN0000072229 for shLacZ, TRCN0000072181 for shGFP, TRCN0000151474 for shATG5, TRCN0000037794 for shVps34#1, and TRC0000296151 for shVps34#2. The miR-338-5p overexpression lentivirus system was purchased from GE Healthcare Dharmacon (Lafayette, CO, USA). Lentiviruses that expressed miR-338-5p or shRNAs were produced according to provider's protocol (http://rnai.genmed.sinica.edu.tw/webContent/web/protocols). The cells were selected using puromycin (P8833; Sigma-Aldrich) (SW480: 15 ng/μl & HCT116: 1 ng/μl).
To establish PIK3C3 overexpression stable cell line, the gene of blasticidin (BSD) was cloned into pCMV-Vps34 plasmid (pCMV-Vps34-BSD) and then transfected into HCT116 cells. The stable cell line was selected by BSD (Cyrusbioscience, Taipei, Taiwan) (5 ng/ul). pTRE2-BSD was used as the control vector.
2.3. Western blotting
Protein expression was measured as previously described [29], using Western blotting with antibodies (Abs) to anti-EIF2C2/AGO2 mouse monoclonal antibody (RN003M, MBL International, Nagoya, Japan), PIK3C3 (#4263; Cell Signaling Technology, Beverly, MA, USA), LC3 (PM036; MBL, Nagoya, Japan), P62 (PM045; MBL), Twist1 (GTX60776; GeneTex, Irvine, CA, USA), Snail (#3879; Cell Signaling Technology), ATG5 (#2630; Cell Signaling Technology), N-cadherin(610,920, BD Transduction Labs, San Diego, CA, USA), E-cadherin (GTX100443; GeneTex), Vimentin (GTX100619; Gentex), Fibronectin (GTX112794; GeneTex) and, β-actin (A5441; Sigma-Aldrich, St. Louis, MO, USA). Full blots were provided in the supplementary information (Supplementary Fig. S6).
Supplementary Fig. S6.
Western full blots were shown. The full blots were equivalent in (a) Fig. 1, (b) Fig. 4, (c) Fig. 5 and (d) supplementary Fig. S4.
2.4. Luciferase assay
To verify whether PIK3C3 was the direct target of miR-338-5p, the predicted target sequence of miR-338-5p within its 3´-UTR was cloned downstream of a luciferase (Luc) gene in the p-miR-reporter Luc plasmid (Ambion; Invitrogen). Cell lysate was assayed using Dual-Glo Luciferase Assay System [E1960] (Promega, Madison, WI, USA) and the results were measured using a luminometer (EG&G Berthold, Wildbad, Germany).
2.5. Ribonucleoprotein-immunoprecipitation (RIP) assay
RIP of miR-338-5p and PIK3C3 was performed in stable miR-338-5p over-expression HCT116 cells by RIP-Assay kit for microRNA (RN1005, MBL). About 4–20 million cells were co-immunoprecipitated with 25 μg of RIP-certified anti-EIF2C2/AGO2 mouse monoclonal antibody (RN003M, MBL) overnight at 4 °C, which was previously conjugated with Sepharose Protein G beads (17-0618-01, GE Healthcare Biosciences, Uppsala, Sweden). Rabbit IgG was used as negative control (RN1005, MBL).
2.6. Injection of tumour cells into spleen
Eight-week-old female NOD/SCID mice were purchased from the NCKU Laboratory Animal Center and maintained in a pathogen-free facility under isothermal conditions with regular photoperiods. The experimental protocol adhered to regulations of the Animal Protection Act of Taiwan and was approved by the NCKU Laboratory Animal Care and User Committee (106127).
Every group of mice (n = 3–4) was anesthetized using intraperitoneally (i.p.) injected Zoletil 50 (25 mg/kg) (Virbac Laboratories, Carros, France) and 2% xylazine (Rompun; Bayer HealthCare, LLC, Leverkusen, Germany). Through a 1- to 2-cm incision on the upper left lateral abdomen, CRC tumour cells (1 × 106) in 100 μl of DMEM were injected into the spleen. After 42 days, the mice were sacrificed and ascites were stained with Liu's stain for tumour cell analysis. The spleen, liver, and lungs were dissected, photographed and weighed. We used a previously described [30] procedure to measure tumour volume.
2.7. Immunohistochemical staining (IHC)
IHC of tissue array was performed using anti-PIK3C3 Ab (#4263; Cell Signaling Technology) as previously described [31]. The resulting complex was labelled with a secondary Ab using a kit (LSAB 2 System; Dako Cytomation). The peroxidase activity was visualized using a diaminobenzidine (DAB) substrate solution (Dako Cytomation) or an aminoethyl carbazole (AEC) substrate solution (Dako Cytomation). The slides were finally counterstained with 10% hematoxylin (Muto Pure Chemicals, Tokyo, Japan).
2.8. Migration and invasion assays
Cell migration in vitro was assessed using a wound-healing analysis at 24 h and a Transwell assay at 48 h (Corning, Corning City, NY, USA). Cell invasion in vitro was estimated using a Transwell assay, in which CRC cells were seeded on Transwell columns coated with a Matrigel membrane (BD Biosciences, San Jose, CA, USA). The cells on the bottom of the membrane were counted after 96 h.
2.9. Immunofluorescent staining
Cells (1 × 105 per well) were seeded on slides and cultured for 48 h. After fixation in formaldehyde for 20 min, slides were treated with 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 30 min. Anti-LC3 polyclonal Ab (PM036; MBL) was incubated at 4 °C overnight and examined under a fluorescent microscope (DP 70; Olympus, Tokyo, Japan).
2.10. Statistical analysis
Parametric test (Student's t-test or ANOVA) was chosen to analyse the data having a normal distribution, and the results presented as means ± standard error of the mean (SEM). For abnormally distributed data or small sample size, non- parametric test (Mann-Whitney U test or Kruskal-Wallis H test) were used. The results are presented as median (interquartile range, IQR). The correlation of miR-338-5p with PIK3C3 mRNA expression was analysed using Spearman tests. Survival analysis was calculated using Log Rank test. Univariate and multivariate associations with overall survival were evaluated using Cox proportional hazards regression models and estimated using the hazard ratio with 95% confidence interval (CI).
3. Results
3.1. PIK3C3 is one of target genes of miR-338-5p
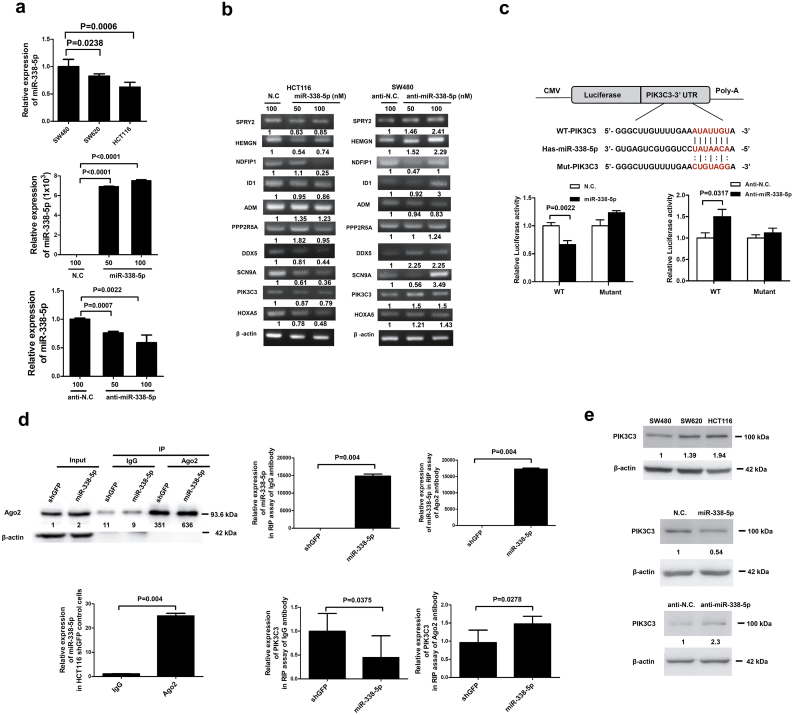
In our pilot study, miR-338-5p was found to associate with metastasis of CRC patients by our in-house miRNA microarray [10]. Then, we performed target gene screening for miR-338-5p [28]. The quest for target genes of miR-338-5p was analysed by TargetScan (http://www.targetscan.org/vert_72/), EBI (https://www.ebi.ac.uk/), and DIANA-microT (http://diana.imis.athena-innovation.gr/DianaTools/index.php) software. Among top 42 consensus target genes, SPRY2, HEMGN, ID1, ADM, DDX5, SCN9A, PIK3C3, and HOXA5 were highly related to CRC (Supplementary table S1). Both NDFIP1 and PPP2R5A were reported as target genes of miR-338-5p in glioblastoma cells [32]. Then, quantitative polymerase chain reaction (QPCR) was performed in CRC cell lines of SW480, SW620 and HCT116 to measure the expression of miR-338-5p. The result showed that expression of miR-338-5p is significantly higher in SW480 cells than in SW620 (P = 0.0238, Mann-Whitney test) and in HCT116 (P = 0.0006, Mann-Whitney test) cells, respectively (Fig. 1a). Thus, SW480 and HCT116 cells were selected for miR-338-5p knockdown and transfection experiments in vitro, respectively. Expression of candidate genes at mRNA level were measured by RT-PCR after HCT116 cells had been transfected with miR-338-5p and SW480 cells with anti-miR-338-5p (inhibitor) (Fig. 1a). Expression of SPRY2, HEMGN, ID1, NDFIP1, DDX5, SCN9A, PIK3C3, and HOXA5 was inhibited in miR-338-5p-overexpressing HCT116 cells. Except for NDFIP1, expression of SPRY2, HEMGN, ID1, DDX5, SCN9A, PIK3C3, and HOXA5 was up-regulated after miR-338-5p had been inhibited (Fig. 1b). On this base, SPRY2, HEMGN, ID1, DDX5, SCN9A, PIK3C3 and HOXA5 may be the actual target genes of miR-338-5p.
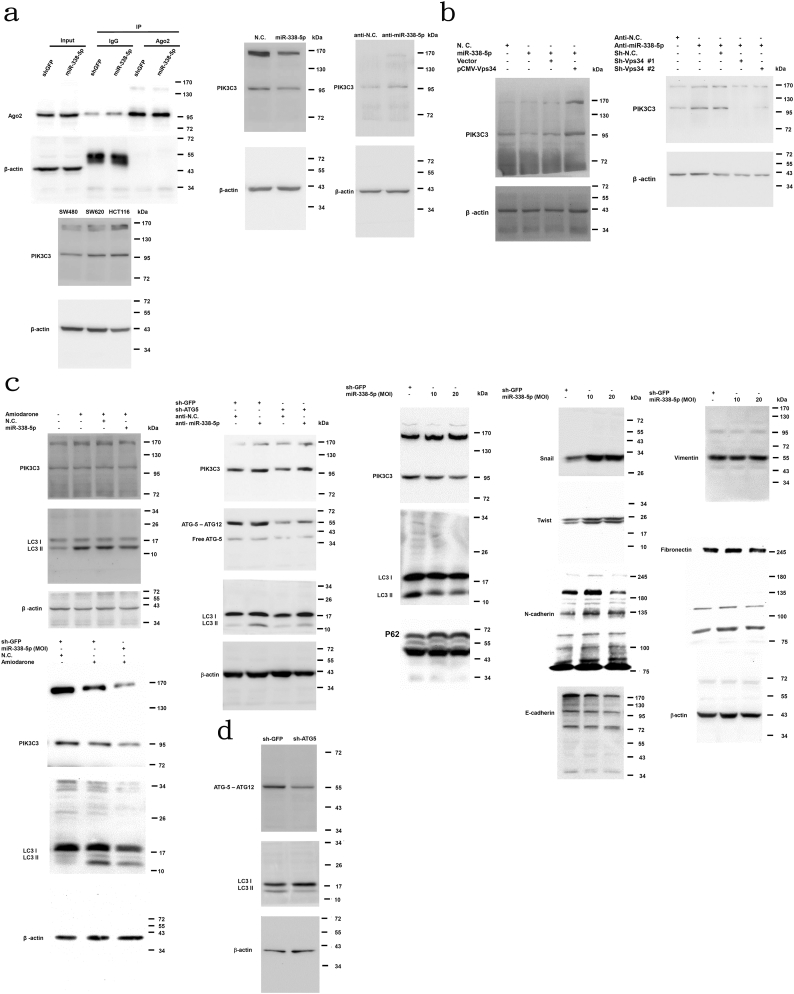
Fig. 1.
PIK3C3 was one of the target genes of miR-338-5p in CRC. (a) QPCR was used to evaluate the miR-338-5p expression in CRC cell lines (SW480, SW620 and HCT116). The data are shown as means ± SEM (n = 5). miR-338-5p (50 nM or 100 nM) or N.C. (100 nM) was transiently transfected into HCT116 cells and analysed using QPCR. The anti-miR- 338-5p (50 nM or 100 nM) or anti-N.C. (100 nM) was transiently transfected into SW480 cells and analysed using QPCR. Data are presented as median (IQR) (n = 3) (P values were analysed using Mann-Whitney tests). (b) RNA expression of miR-338-5p target genes (SPRY2, HEMGN, NDFIP1, ID1, ADM, PPP2R5A, DDX5, SCN9A, PIK3C3, and HOXA5) was measured using RT-PCR after transfection of miR-338-5p or anti-miR-338-5p into SW480 cells. (c) The target sequence of PIK3C3 was constructed into 3´-UTR of p-miR-reporter luciferase plasmid downstream of luciferase gene. Wild-type (WT) and mutant-type (mutant) target sequences are shown. The SW480 cells were transfected with WT or mutant p-miR-report-PIK3C3 plasmid (5 μg/ml) and co-transfected with miR-338-5p, anti-miR-338-5p, N.C. or anti-N.C. (100 nM), respectively. Data are presented as means ± SEM (n = 5) (P values were analysed using Mann-Whitney tests). (d) RIP assay was performed to precipitate the Ago2 complexe from stable miR-338-5p overexpression or shGFP control cells. Western blot was used to confirm the quality of RIP. Expression of miR-338-5p and PIK3C3 RNA in Ago2 RIP fractions was measured by QPCR assay. Data are presented as median (IQR) (n = 5) (P values were analysed using Mann-Whitney tests). (e) Expression of PIK3C3 protein in CRC cell lines was evaluated using Western blotting. miR-338-5p or its N.C. was transiently transfected into HCT116 cells, and anti-miR-338-5p or anti-N.C. was transiently transfected into SW480 cells. Expression of PIK3C3 protein was assessed using Western blotting. β-actin was used as an internal control.
We showed that PIK3C3 is the most important gene related to autophagy (supplementary table S1) and was reported to induce autophagy [21]. Prior studies reported down-regulated autophagy-related genes (ATG5 and Beclin1) in primary CRC [33,34] and their association with poor patient prognosis [34,35]. Thus, we speculated that PIK3C3 is the most plausible target gene of miR-338-5p. To investigate its involvement in miR-338-5p- mediated biological effects, wild-type (wt) and mutant-type (mt) PIK3C3 3´-UTR target sequences were constructed to investigate whether PIK3C3 is one of target genes of miR-338-5p. A significantly (P = 0.0022, Mann-Whitney test) lower luciferase activity of p-MIR-PIK3C3 was observed after both miR-338-5p and wt-p-MIR-PIK3C3 had been transfected into SW480 cells. In contrast, significantly (P = 0.0317, Mann-Whitney test) higher p-MIR-PIK3C3 luciferase activity was demonstrated after miR-338-5p inhibitor had been transfected into SW480 cells. However, the regulatory effect was abolished when p-MIR-mt-PIK3C3 luciferase reporter plasmid was transfected into SW480 cells (Fig. 1c). The results support that miR-338-5p is targeting to 3’UTR of PI3KC3.
To confirm this hypothesis, stable miR-338-5p over-expression and shGFP control HCT116 cell lines were established for RIP assay. Protein argonaute-2 (Ago2) is the core of RNAi-induced silencing complex (RISC). In RNAi-pathway, RISC binds to microRNA and its target-gene, resulting in either mRNA degradation or translational repression [36]. Compared with anti-IgG, Ago2 antibody specifically precipitated with ago2 and miR-338-5p in shGFP control cell line (P = 0.004, Mann-Whitney test), indicating that miR-338-5p bound to Ago2 (Fig. 1d). When IgG or Ago2 was precipitated, expression of miR-338-5p was higher in miR-338-5p over-expression cells than in shGFP control cells (P = 0.004, Mann-Whitney test). Compared with shGFP control cells, lower level of PIK3C3 RNA was observed in RIP-IgG lysate of miR-338-5p over-expression cells (P = 0.0375, Mann-Whitney test), supporting that miR-338-5p suppresses the expression of PIK3C3 RNA. In contrast, higher level of PIK3C3RNA was demonstrated in RIP-Ago2 lysate of stable miR-338-5p overexpression HCT116 cells. These results confirm that PIK3C3 RNA binds to miR-338-5p (Fig. 1d).
The SW480 cells had the lowest level of PIK3C3 protein expression, while HCT116 cells had the highest level. Expression of PIK3C3 was negatively correlated with that of miR-338-5p (Fig. 1a and e). Moreover, expression of PIK3C3 in HCT116 cells was inhibited after transfection of miR-338-5p. In contrast, PIK3C3 expression was higher in SW480 cells when anti-miR-338-5p was transfected (Fig. 1e). All of results described above support for PIK3C3 as one of the target genes of miR-338-5p in CRC.
3.2. Correlation of miR-338-5p with PIK3C3 expression and its clinical implication
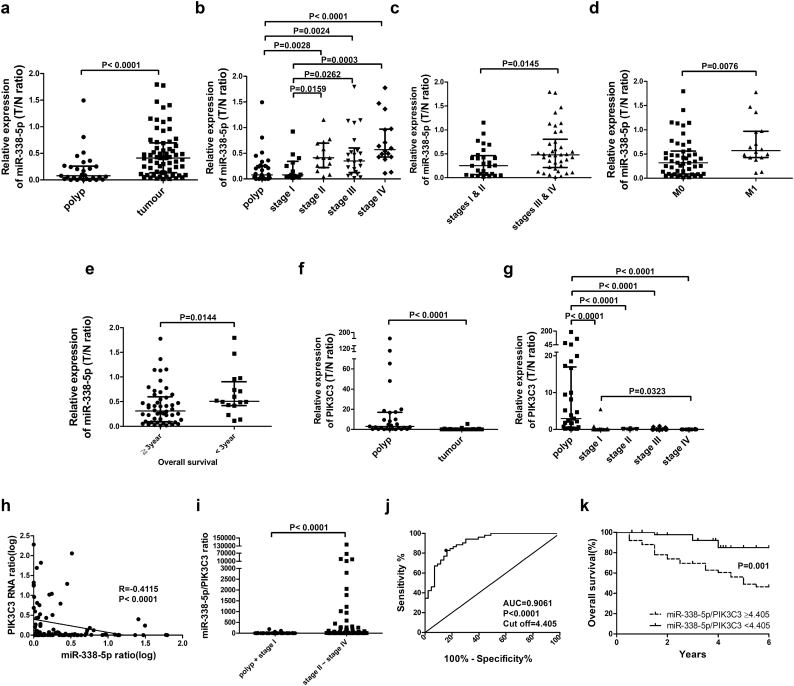
The association of miR-338-5p expression with clinicopathologic indicators and patient outcome was analysed in an independent cohort of CRC tumours (n = 66) and colorectal polyps (n = 29). MiR-338-5p expression was measured using real-time PCR. A significantly lower miR-338-5p ratio (tumour/adjacent normal, T/N) was demonstrated in benign polyps than in CRCs (Fig. 2a) (Supplementary table S2). In early stages of colorectal neoplasia (polyps or stage I CRC), expression of miR-338-5p was lower than that of late stages CRC (stage II- IV) (Fig. 2b). In addition, overexpression of miR-338-5p was positively related to tumour staging (P = 0.0019, Kruskal-Wallis test) (Supplementary table S3), advanced tumours (stage I-II vs. stage III-IV, P = 0.0145, Mann-Whitney test) (Fig. 2c), distant metastasis (P = 0.0076, Mann-Whitney test) (Fig. 2d), and overall survival (P = 0.0144, Mann-Whitney test) (Fig. 2e) (Supplementary table S3), respectively. Then, expression of PIK3C3 RNA was measured in colorectal polyps and CRC tissue using real-time PCR and correlated with clinicopathologic indicators and patient outcome (Supplementary tables S4 and S5). The ratio of PIK3C3 (T/N) was significantly higher in benign polyps than in CRC (stages I–IV) (Fig. 2f and g) (Supplementary table S4). These findings support the significance of miR-338-5p and PIK3C3 in the progression of CRC tumourigenesis.
Fig. 2.
The association of miR-338-5p and PIK3C3 with tumour staging and patient survival. Expression of miR-338-5p or PIK3C3 mRNA was assayed using real-time QPCR. (a) Expression of miR-338-5p (T/N ratio) was calculated in benign polyps and tumour tissue of CRC (n = 95). (b) Expression of miR-338-5p (the ratio of T/N) was calculated in benign polyps and CRC specimens cross different stages (n = 95). Expression of miR-338-5p (T/N ratio) was calculated and compared in relation to stage I, II vs. stage III, IV (c), M0 vs. M1 (d) and overall survival (e) of CRC (n = 66). (f) RNA expression of PIK3C3 (T/N ratio) was calculated in benign polyps and tumour tissue of CRC (n = 95). (g) RNA expression of PIK3C3 (T/N ratio) was calculated in benign polyps and CRC specimens cross all stages (n = 95). Data are presented as median (IQR) (P values were analysed using Mann-Whitney test). (h) The correlation of miR-338-5p with PIK3C3 mRNA expression (the ratio of T/N) was estimated in polyps and CRC specimens (n = 95) using linear regression (Data were analysed by Spearman tests). (i) The miR-338-5p/PIK3C3 ratio was calculated in benign polyps, early stage (stage I) and late-stages (stages II-IV) of CRC specimens (n = 95). Data are presented as median (IQR) (P values were analysed using Mann-Whitney tests). (j) AUCs of ROC curves, cutoff values, specificity, and sensitivity are shown for benign polyps and for stage I and stage II-IV CRC specimens. An AUC between 0.5 and 0.6 was defined as nondiscriminatory, between 0.6 and 0.7 as acceptable, between 0.7 and 0.8 as excellent, and between 0.8 and 0.9 as outstanding (n = 95). (k) Kaplan-Meier analysis was used to estimate the overall survival of polyps and CRC patients (n = 95) at 6 years after surgery (P values were analysed using Log Rank tests). The cut-off of miR-338-5p/PIK3C3 ratio set at 4.405 was a practical prognostic biomarker for CRC patients.
Expression of PIK3C3 was inversely related to miR-338-5p. Linear regression analysis showed that PIK3C3 expression was inversely correlated with miR-338-5p in vivo (P < 0.0001, r = −0.4115, Spearman test) (Fig. 2h). Late-stage tumours (stages II - IV) had significantly (P < 0.0001, Mann-Whitney test) higher miR-338-5p/PIK3C3 ratios than did early-stage colorectal neoplasia (Fig. 2i). The miR-338-5p/PIK3C3 ratio successfully discriminated tumour grading with area under the curve (AUC) value estimated at 0.9061 and cut-off set at 4.405 (Fig. 2j). The miR-338-5p/PIK3C3 ratios ≥4.405 significantly predicted poor overall survival (P = 0.001, Log Rank test) of patients (Fig. 2k). In univariate analysis, polyps or CRCs with miR-338-5p/PIK3C3 ratio ≥ 4.405 had 5.418 times higher risk of patient death than those with miR-338-5p/PIK3C3 ratio < 4.405 (P = 0.002, Cox proportional hazards regression model). patients with stages II–IV CRC had 20.908 times higher risk of patient death than that of polyps or stage I CRC (P = 0.003, Cox proportional hazards regression model) (Supplementary table S6). In multivariate analysis, patients with stages II–IV CRC had 13.921 times higher risk of patient death than those with polyps or stage I CRC (P = 0.014, Cox proportional hazards regression model). No prognostic significance for miR-338-5p/PIK3C3 ratio was observed in the multivariate model (Supplementary table S7). Taken together, miR-338-5p is inversely correlated with PIK3C3 expression and plays an important role in the progression of colorectal cancer. The miR-338-5p/PIK3C3 ratio is a promising prognostic biomarker in the selection of CRC patients who may require aggressive treatment strategy.
3.3. Inhibited PIK3C3 expression in miR-338-5p-treated cells stimulates CRC metastasis
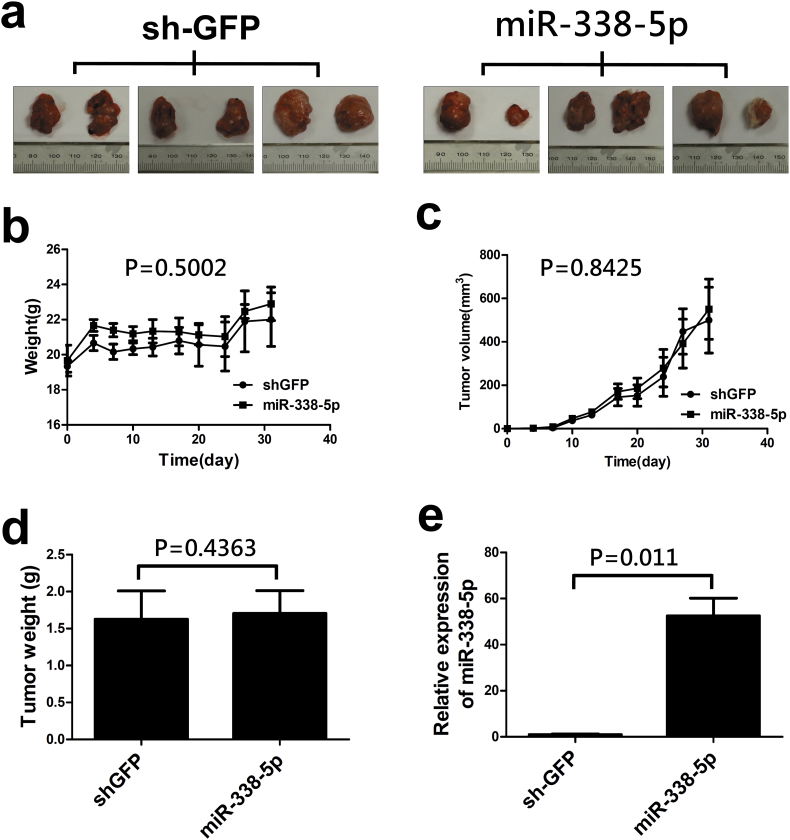
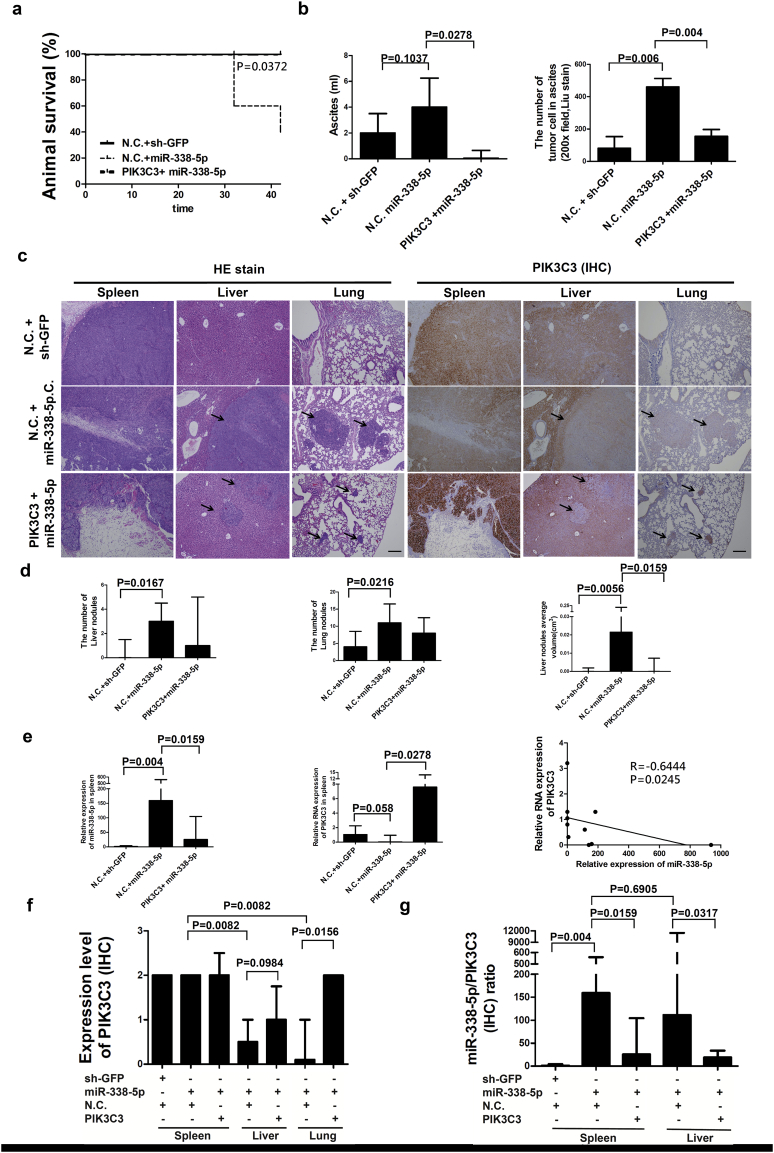
To clarify the involvement of miR-338-5p in the progression of CRC in vivo, stable miR-338-5p-overexpression and PIK3C3-overexpression cell lines were established in HCT116 cells, respectively. A xenograft mouse model was established to verify the tumourigenic potential of miR-338-5p in vivo. Overexpression of miR-338-5p did not affect tumour weight or volume (Supplementary Fig. S1). Stable miR-338-5p cells, miR-338-5p and PIK3C3 co-overexpression cells, and shGFP control cells were injected into the spleen of NOD/SCID mice to examine their potential impact on CRC metastasis (Supplementary Fig. S2a). Mice injected with overexpression of miR-338-5p had a lower survival (P = 0.0372, Log Rank test). However, when PIK3C3 was activated in stable miR-338-5p overexpression cells, survival rate of mice was recovered (Fig. 3a). Overexpression of miR-338-5p did not affect tumour growth in the spleen. When PIK3C3 was overexpressed, volume of miR-338-5p overexpression tumours in the spleen was suppressed (Supplementary Fig. S2a, b, c). The results support that overexpression of miR-338-5p negatively modulates survival of mice through suppression of PIK3C3, and PIK3C3 could inhibit tumour growth in vivo.
Supplementary Fig. S1.
The effect of miR-338-5p was on CRC growth in a mouse xenograft model. HCT116 cells infected with miR-338-5p or shGFP lentivirus were injected into both sides of back in the NOD/SCID mice. (a) Tumours were dissected out and photographed on day 31. The kinetics of mouse body weight (b) and tumour volume (c) were monitored during study period (P value was analysed using ANOVA). (d) The tumour weight was measured when mice were sacrificed. (e) The tumours were assayed for miR-338-5p RNA expression using QPCR. Data are presented as median (IQR) (n = 6) (P value was analysed using Mann-Whitney tests).
Fig. 3.
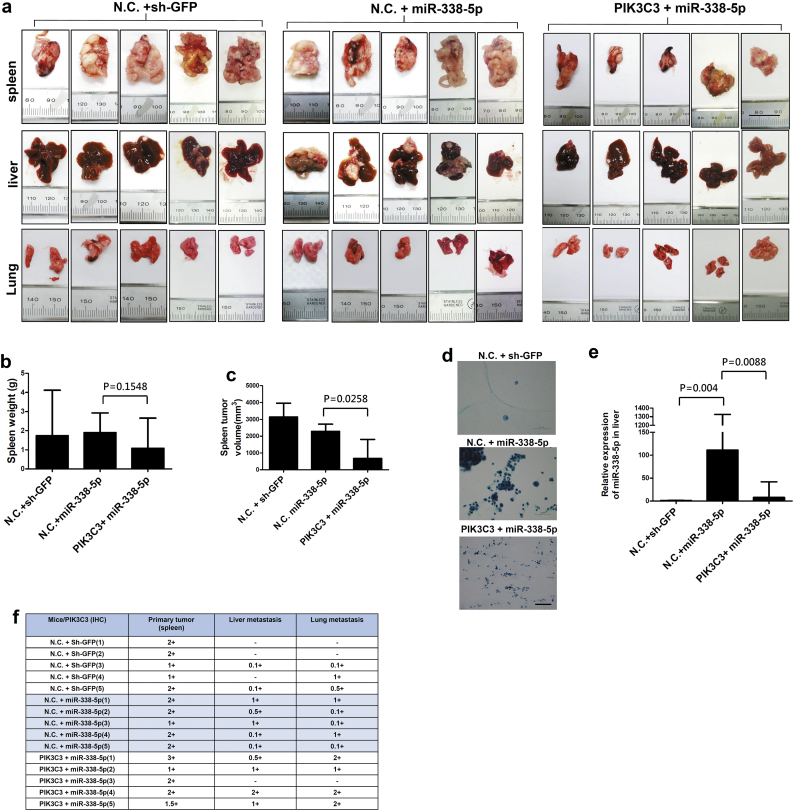
Inhibition of PIK3C3 in miR-338-5p stable cells promoted CRC metastasis in vivo. HCT116 stable cells were injected into the spleens of NOD and SCID mice. After 42 days, the spleen, liver, and lung were dissected out. (a) Kaplan-Meier analysis was used to calculate the overall survival rate of mice (P values were analysed using Log Rank tests). (b) The amount of ascites was measured and tumour cells in ascites were demonstrated by Liu stain. Data are presented as median (IQR) (n = 5) (P values were analysed using Mann-Whitney tests). (c) Tumours in the spleen, liver, and lung were examined by hematoxylin and eosin (H&E) stain and PIK3C3 IHC staining (100×). The metastatic tumours were indicated by arrow point. IHC showed that PIK3C3 was high expression in primary tumour and metastatic nodules, when PIK3C3 was overexpressed. (d) The number and volume of metastatic tumours were measured in the liver and lung, respectively. The scale bar = 200 μm. Data are presented as median (IQR) (n = 5) (P values were analysed using Mann-Whitney tests). (e) Expression of miR-338-5p or PIK3C3 RNA in primary tumour of spleen was assayed by QPCR. Data are expressed as median (IQR) (n = 5) (P values were analysed using Mann-Whitney tests). IHC of PIK3C3 expression (f) and ratio of miR-338-5p/PIK3C3 (g) were measured in the splenic primary tumour, metastasis in the liver and lung, respectively. Data are expressed as median (IQR) (n = 5) (P values were analysed using Mann-Whitney tests). Correlation was calculated using linear regression (Data were analysed by Spearman tests).
Besides, more tumour cells with overexpression of miR-338-5p were observed in the ascites of mice (P = 0.006, Mann-Whitney test) than shGFP control. When PIK3C3 was overexpressed in miR-338-5p stable cells, both quantity of ascites (P = 0.006, Mann-Whitney test) and number of tumour cell in the ascites (P = 0.004, Mann-Whitney test) were suppressed (Fig. 3b and Supplementary Fig. S2d). It appears that miR-338-5p may promote peritoneal metastasis of CRC and the effect could be reversed by PIK3C3. Significantly more metastatic nodules were observed in the livers (P = 0.0167, Mann-Whitney test) and lungs (P = 0.0216, Mann-Whitney test) of mice after injection with stable miR-338-5p cells (Fig. 3c and d). Furthermore, overexpression of miR-338-5p promoted the growth of liver nodules (P = 0.0056, Mann-Whitney test), while overexpression of PIK3C3 in miR-338-5p stable cells inhibited the growth of metastatic tumours (P = 0.0159, Mann-Whitney test) in the liver (Fig. 3d).
In primary xenograft tumours, expression of miR-338-5p RNA was negatively associated with PIK3C3 levels (P = 0.0245, R = −0.6444, Spearman test) when miR-338-5p was overexpressed (Fig. 3e). IHC showed that PIK3C3 protein expression is significantly higher in tumours with miR-338-5p overexpression in the spleen than that of metastatic tumours in the liver (P = 0.0082, Mann-Whitney test) and lung (P = 0.0082, Mann-Whitney test), respectively (Fig. 3c and f). Compared with miR-338-5p stable cells, PIK3C3 expression was higher in the metastatic tumours to the liver (P = 0.0984, Mann-Whitney test) and lung (P = 0.0156, Mann-Whitney test) with co-expressed miR-338-5p and PIK3C3 stable cells, respectively (Fig. 3c, f and Supplementary Fig. S2f). As with colon cancer patients, miR-338-5p/PIK3C3 ratio was higher in primary tumours with miR-338-5p overexpression than control (P = 0.004, Mann-Whitney test) (Fig. 3g). Overexpression of PIK3C3 reversed the miR-338-5p/PIK3C3 ratio in primary tumours (P = 0.0159, Mann-Whitney test) and metastatic liver nodules (P = 0.0317, Mann-Whitney test) (Fig. 3g). Liver metastatic tumours showed a trend toward higher miR-338-5p/PIK3C3 ratio than primary tumours in the spleen, although no statistical significance was reached (P = 0.6905, Mann-Whitney test) (Fig. 3g). Together, miR-338-5p involves in metastasis of CRC in vivo through silencing of PIK3C3, while growth of metastatic tumour is inhibited by PIK3C3.
3.4. MiR-338-5p induces migration and invasion of CRC through PIK3C3
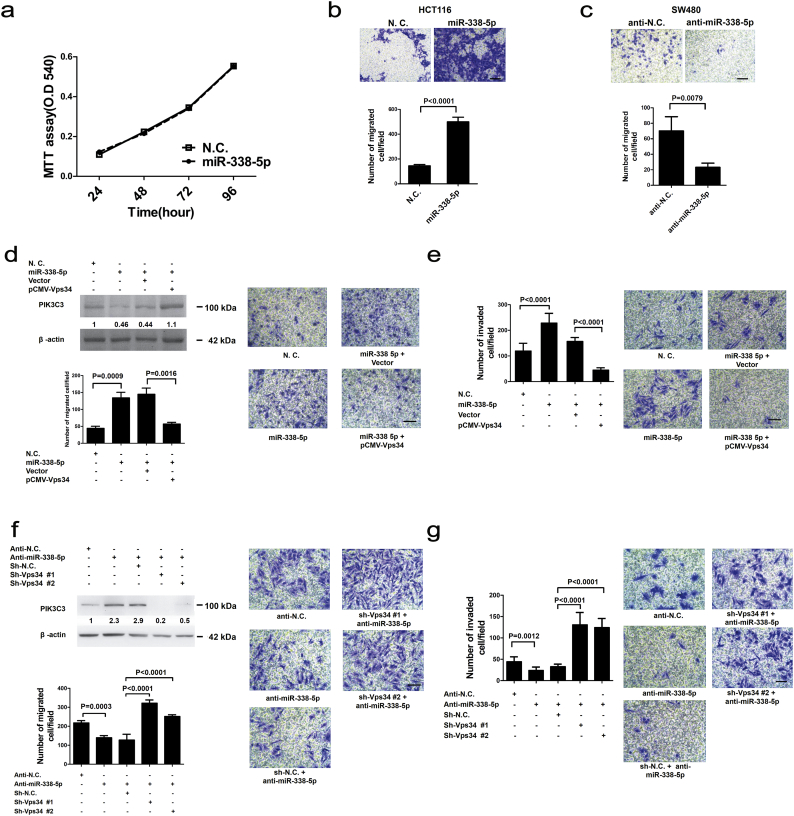
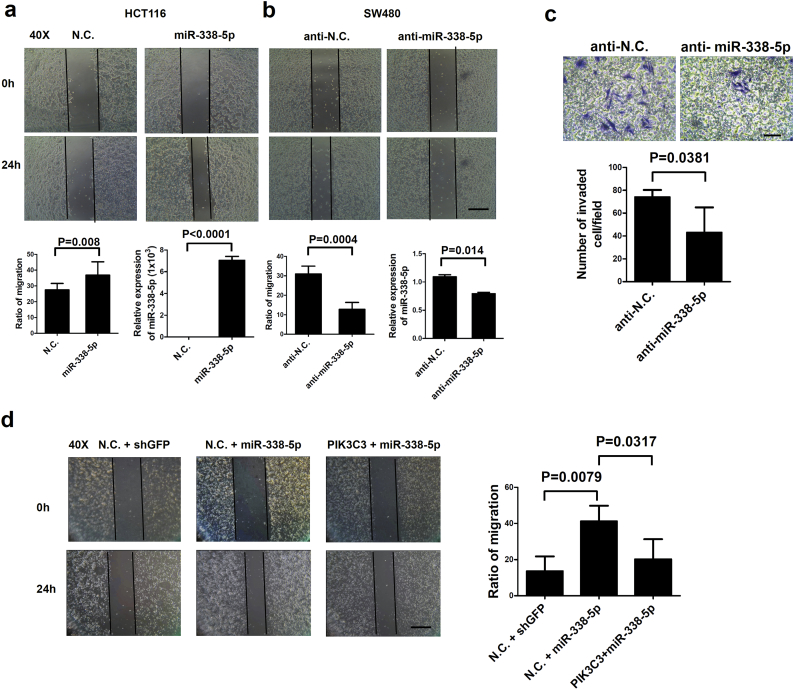
To examine the biological effects of miR-338-5p overexpression in vitro, methylthiazol tetrazolium (MTT) assay did not demonstrate altered growth rate of HCT116 cells that had been transfected with miR-338-5p (Fig. 4a). Both wound healing and transwell migration assays showed that cell migration is significantly higher in miR-338-5p-transfected HCT116 cells (Fig. 4b; Supplementary Fig. S3a). In contrast, cell migration was significantly inhibited in anti-miR-338-5p-transfected SW480 cells (Fig. 4c; Supplementary Fig. S3b). Transwell invasion capability was significantly inhibited in SW480 cells that had been transfected with anti-miR-338-5p (Supplementary Fig. S3c). Thus, miR-338-5p can promote CRC migration and invasion in vitro without noticeable effect on cell proliferation.
Fig. 4.
Involvement of PIK3C3 in miR-338-5p mediated cell migration and invasion. (a) miR-338-5p or negative control (N.C.) (100 nM) was transiently transfected into HCT116 cells, put into 96-well plates (4000/well) and cultured for 24 h. The number of cells was calculated every 24 h using MTT assays for four consecutive days. Data are expressed as means (n = 6) (P values were analysed using ANOVA). (b, c) Cell migration was analysed using transwell migration assays 48 h after transfection with miR-338-5p or anti-miR-338-5p into HCT116 or SW480. The transfected cells were seeded on transwell plates, and migrated cells at the bottom of membrane were calculated. The scale bar represents 50 μm. The number of migrated HCT116 cells are shown as means ± SEM (n = 8) (P values were analysed using t tests).The number of migrated SW480 cells are shown as median (IQR) (n = 5) (P values were analysed using Mann-Whitney tests). (d) miR-338-5p or pCMV-Vps34 plasmid was transiently transfected into SW480 cells and analysed for PIK3C3 protein expression 48 h later. Cell migration was evaluated using transwell assay. Forty-eight hours after transfection, SW480 cells were seeded on transwell plates, and those cells appeared at the bottom of membrane were counted. Data are expressed as means ± SEM (n = 8) (P value was analysed using t tests). (e) Both miR-338-5p and pCMV-Vps34 plasmids were transiently transfected into SW480 cells. Transwell columns were coated with a Matrigel membrane and seeded with transfected cells 48 h later. The invaded cells at the bottom of membrane were counted after 96 h. Data are expressed as means ± SEM (n = 8) (P value was analysed using t tests). (f) SW480 cells were transiently transfected with anti-miR-338-5p (100 nM) and shVps34 lentivirus and assayed for PIK3C3 protein expression. Migration of SW480 cells was assayed using transwell assay. Number of migrated CRC cells was counted after 48 h. Data are expressed as means ± SEM (n = 8) (P value was analysed using t tests). (g) Transwell invasion assays were used to analyse SW480 cells transfected with anti-miR-338-5p or shVps34 lentivirus and counted for invaded cells after 96 h. Data are expressed as means ± SEM (n = 8) (P value was analysed using t tests). The scale bar represents 50 μm.
Supplementary Fig S3.
The effect of miR-338-5p was on migration and invasion. CRC cell migration and invasion were measured using wound-healing and transwell assays, respectively. (a) The miR-338-5p or N.C. (100 nM) was transiently transfected into HCT116 cells. Twenty-four hours after creation of wounding, images of wound-healing were recorded using an optical microscope (40×). Expression of miR-338-5p was confirmed using QPCR. The width between wounding was calculated at each time point and expressed as a percentage relative to that of time zero. (b) Anti-miR-338-5p or anti-negative control (anti-N.C.) (100 nM) was transiently transfected into SW480 cells. Twenty-four hours after wounding, images of wound-healing were recorded using an optical microscope (40×). The scale bar represents 100 μm. The ratio of migration and expression of miR-338-5p are shown as median (IQR) (n = 5) (P value was analysed using Mann-Whitney tests). (c) Cell invasion was analysed using transwell invasion assays 96 h after transient transfection of anti-miR-338-5p (100 nM) into SW480 cells. The transwell columns were coated with a Matrigel membrane. The transfected cells were seeded on the transwell plates. Only invaded cells at the bottom of membrane after 96 h were counted. The scale bar represents 50 μm. Data are shown as midian(IQR) (n = 5) (P value was analysed using Mann-Whitney tests). (d) Wound healing assays were used to analyse PIK3C3 over expression stable HCT116 cells infected with miR-338-5p or shGFP lentivirus and counted for migrated cells after 24 h. The scale bar represents 100 μm. Data are expressed as median (IQR) (n = 4–5) (P value was analysed using Mann-Whitney tests).
To clarify the involvement of PIK3C3 in miR-338-5p-related biological effects, pCMV-Vps34 (PIK3C3) vector was prepared for experiments in vitro. Migration (P = 0.0009, t test) and invasion (P < 0.0001, t test) were significantly induced in SW480 cells overexpressing miR-338-5p than in SW480 cells transfected with control vector. In contrast, cell migration and invasion were significantly (P = 0.0016, t test) inhibited when PIK3C3 was reactivated (Fig. 4d and e). The conclusion could be confirmed by wound healing assay using stable miR-338-5p and PIK3C3 over-expression HCT116 cells. Overexpression of PIK3C3 rescued the inhibition of cell migration by overexpressing miR-338-5p (Supplementary Fig. S3d). Moreover, lentivirus for shVps34 (PIK3C3) experiment was prepared from reverse side. PIK3C3 expression was higher and cell migration (P = 0.0003, t test) and invasion (P = 0.0003, t test) were significantly lower in SW480 cells after transfection of anti-miR-338-5p. But, PIK3C3 expression was lower and cell migration and invasion were rescued after transfection with shVps34 lentivirus (Fig. 4f and g). The results support that miR-338-5p stimulates CRC migration and invasion through inhibition of PIK3C3.
3.5. The autophagy pathway involves in CRC migration and invasion induced by miR-338-5p
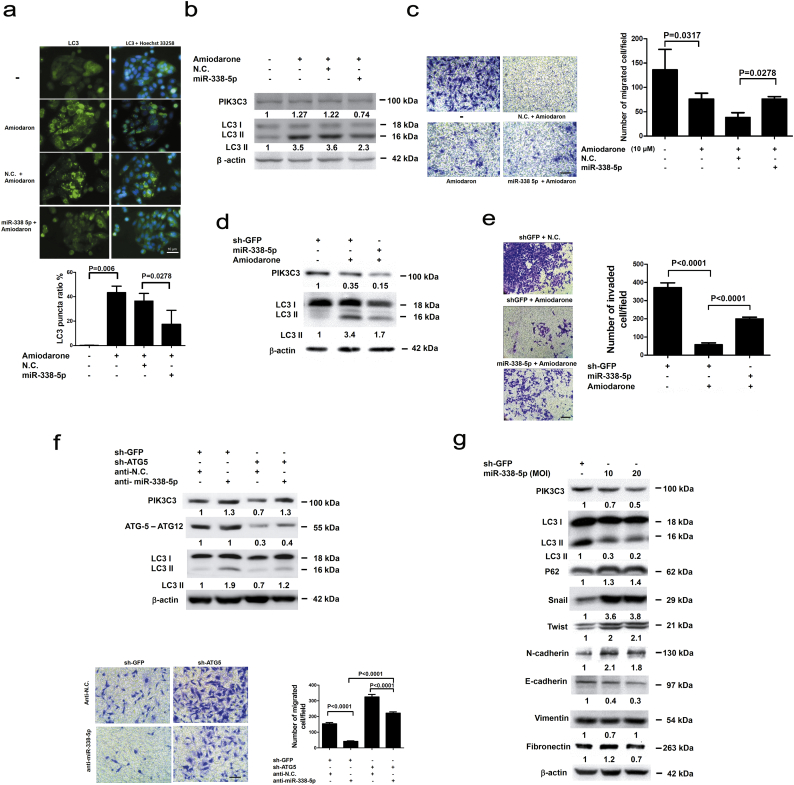
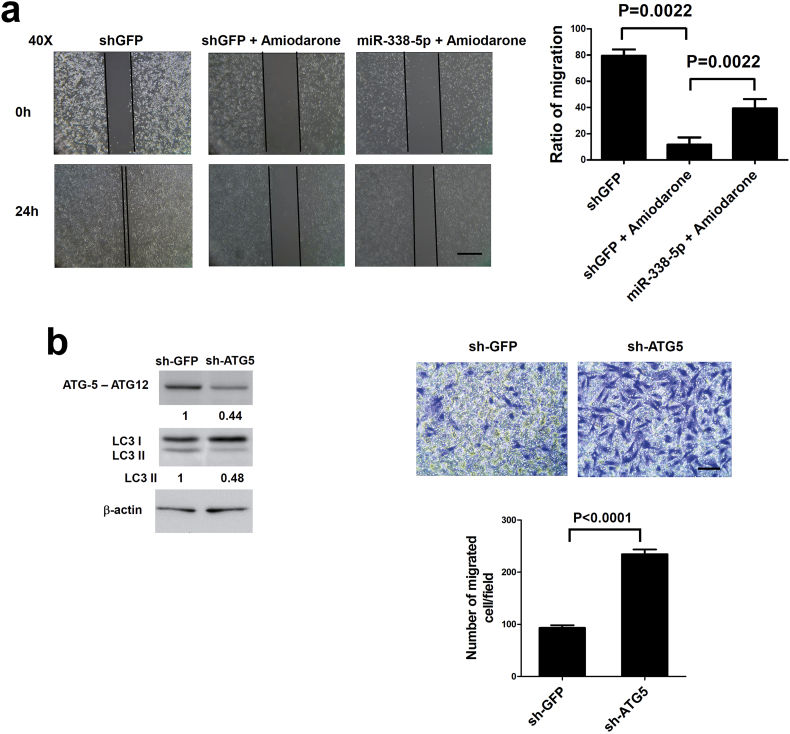
Because PIK3C3 was related to autophagy, the involvement of autophagy in miR-338-5p induced cell migration was assessed. The light chain 3 (LC3) puncta (microtubule associated protein 1A/1B-LC3) formation and LC3 type II (LC3-II) protein expression was examined in cells treated with amiodarone (an autophagy inducer) by immunofluorescent assay [37,38]). Significant (P = 0.006, Mann-Whitney test) LC3 puncta formation (Fig. 5a) and LC3-II expression (Fig. 5b and d) were detected in CRC cells treated with amiodarone (10 μM). When PIK3C3 was suppressed, amiodarone-induced autophagy was significantly inhibited in miR-338-5p-overexpressing cells (Fig. 5a, b, d). Cell migration (Fig. 5c, Supplementary Fig. S4a) and invasion (Fig. 5e) were also significantly inhibited in amiodarone-treated cells, but could be rescued in miR-338-5p-overexpressing cells. These findings indicate that miR-338-5p increases migration and invasion of CRC cells through repression of autophagy.
Fig. 5.
Involvement of autophagy or EMT pathway in the miR-338-5p-related CRC migration in vitro. (a) LC3 puncta were labelled with anti-LC3 antibody and fluorescent isothiocyanate-conjugated goat anti-rabbit IgG (green fluorescence). SW480 cell nuclei were stained with Hoechst 33258 (blue fluorescence). Images were photographed using florescent microscopy (the scale bar represents 10 μm). Data are presented as median (IQR) (n = 5) (P value was analysed using Mann-Whitney tests). (b) SW480 cells were treated with 10 μM of amiodarone for 48 h and then transfected with 100 nM of miR-338-5p or N.C. PIK3C3 and LC3 expression was measured using Western blot. (c) For transwell assay, SW480 cells were first treated with 10 μM of amiodaron and then transfected with/without 100 nM of miR-338-5p. Data are presented as median (IQR) (n = 5) (P value was analysed using Mann-Whitney tests).The scale bar represents 50 μm. (d) After infection with lentivirus carrying miR-338-5p or sh-GFP, HCT116 stable cells were treated with amiodarone (10 μM) for 48 h. Expression of PIK3C3 and LC3 was assayed by Western blot. (e) Transwell invasion assay was performed on HCT116 cells after infection with miR-338-5p or shGFP lentivirus and counted for invaded cells for 48 h. The scale bar represents 50 μm. Data are expressed as means ± SEM (n = 8) (P value was analysed using t tests). (f) SW480 cells were infected with sh-ATG-5 lentivirus and then transfected with 100 nM of miR-338-5p or N.C. Expression of ATG5-ATG12 and LC3 was measured using Western blot. Representative results of transwell assays. The scale bar represents 50 μm. The number of migrated cells are shown as means ± SEM (n = 8) (P values were analysed using t tests). (g) After infection with lentivirus carrying miR-338-5p, western blot was used to analyse the expression of PIK3C3, LC3, p62, Snail, Twist, N-cadherin, E-cadherin, vimentin, fibronectin and β-actin, respectively, in HCT116 cells. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To confirm this observation, a shATG5 lentivirus was used to modulate autophagy in vitro. The autophagy activity (LC3 II) was inhibited in ATG5 knockdown SW480 cells (Supplementary Fig. S4b) in association with increased cell migration (Supplementary Fig. S4b). When SW480 cells were transiently transfected with anti-miR-338-5p (miR-338-5p inhibitor), PIK3C3 and LC3-II was expressed and cell migration was significantly (P < 0.0001, t test) inhibited. Suppression of autophagy through silencing of ATG5 reversed the inhibited migration of SW480 cells transfected with anti-miR-338-5p (P < 0.0001, t test), However, migration ability was lower than that of receiving sh-ATG5 lenti-virus infection (P < 0.0001, t test) (Fig. 5f), implying the existence of miR-338-5p independent mechanism(s) involved in autophagy related cell migration. We speculate that miR-338-5p stimulates migration and invasion of CRC cells in part through inhibition of autophagy.
To validate our hypothesis, stable miR-338-5p-overexpression HCT116 cell line was analysed by western blot. When miR-338-5p was overexpressed, both PIK3C3 and LC3 II protein expression were inhibited together with increased p62 (SQSTM1, an autophagosome degradation marker [39]). In addition, EMT phenotype, as demonstrated by down-regulated E-cadherin, and accumulation of N-cadherin, Snail and Twist proteins, was induced (Fig. 5g). However, expression of vimentin and fibronectin was not affected (Fig. 5g). The results implied that miR-338-5p regulates EMT in conjunction with inhibition of autophagy. Altogether, PIK3C3-related autophagy pathway explains part of miR-338-5p-mediated CRC migration, invasion and metastasis in vitro.
4. Discussion
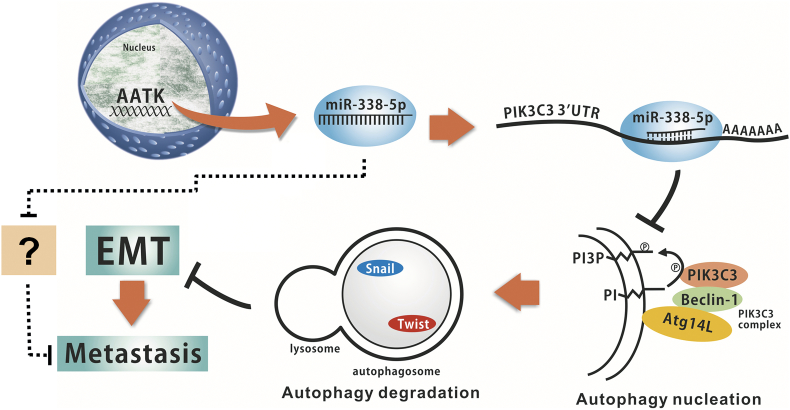
In this model experiment, expression of miR-338-5p induces CRC migration and invasion in vitro and metastasis in vivo by inhibiting PIK3C3. Moreover, miR-338-5p inhibits autophagy, suppresses the degradation of snail and twist and promotes tumour metastasis through EMT. However, suppression of PIK3C3 could only explain part of inhibitory effect of autophagy on miR-338-5p. The fact that autophagy could not completely reverse the miR-338-5p related migration suggest the existence of additional molecular mechanisms (Supplementary Fig. S5). Actually, screening of miR-338-5p candidate genes discovered the potential of extra paths, e.g., SPRY2, NDFIP1, and DDX5, in the pathogenesis of miR-338-5p-mediated biological effects. On the contrary, CRC cell migration did not completely recover when miR-338-5p was inhibited and ATG5 knocked down, implying the existence of miR-338-5p-independent mechanisms in the autophagy-mediated tumourigenesis. Further studies are required to clarify that diverse biological effects of microRNAs in the pathogenesis of CRC are regulated by multiple mechanisms in vivo.
Supplementary Fig. S5.
Hypothetical model of miR-338-5p involved in metastatic progression of human colorectal cancer. MiR-338-5p is located at an intronic region within AATK gene. MiR-338-5p cleaves PIK3C3 mRNA and inhibits the membrane nucleation of autophagosome by phosphorylation of phosphatidylinositol (PI). Inhibited autophagy then suppresses the degradation of Snail and Twist and promotes the tumour metastasis through EMT pathway. But, autophagy could not reverse this pathway completely, indicating that miR-338-5p may promote metastasis of CRC by inhibiting other target genes.
We further demonstrated that miR-338-5p is positively associated with advanced tumour stages, metastasis, and poor-overall survival in CRC patients. Thus, a recent report [40] of the metastasis-specific miRNA signature of CRC can perhaps be supplemented by miR-338-5p. Together with increased serum levels of miR-338-5p in the advanced stages of CRC [16], miR-338-5p expression might be a practical prognostic biomarker in CRC patients. Analysis of clinical cohort showed that miR-338-5p is inversely related to PIK3C3 expression and high miR-338-5p/PIK3C3 ratio is correlated with poor patient survival. Bilegsaikhhan et al. reported that serum miR-338-5p is a potential non-invasive diagnostic biomarker for detecting CRC [17]. In addition to CRC, up-regulated miR-338 or miR-338-5p was also observed in various human cancers [[41], [42], [43], [44]], highlighting potential multiple origins of miR-338-5p in the serum. Therefore, serum biomarker assessment, such miR-338-5p, is not specific to CRC.
We provide evidence that miR-338-5p/PIK3C3 ratio in the CRC tissue is positively associated with tumour staging and patient survival. This is the first report showing the implication of miR-338-5p/PIK3C3 axis in the metastatic progression of CRC in vitro and in vivo. Therefore, miR-338-5p/PIK3C3 ratio is a promising prognostic biomarker for CRC patients who require aggressive treatments. Moreover, PIK3C3 is a novel therapeutic target in CRC.
Currently, relatively little information is available regarding PIK3C3 in human cancer. We showed that PIK3C3 functions as a suppressor in CRC. This designation is in contrast to bladder cancer, HCC, and renal cell carcinoma [[45], [46], [47]], in which autophagy mediated by PIK3C3 complex was reported to promote tumour growth. In this situation, SAR405, a low-molecular mass kinase inhibitor of PIK3C3 [47], and PIK3C3 inhibitor MPT0L145 [45] warrant investigation as targeting agents. Interestingly, overexpression of PIK3C3 also inhibited the expression of miR-338-5p in primary splenic tumours of miR-338-5p stable cells (Fig. 3E) and liver nodules (Supplementary Fig. S2E). Even though PIK3C3 may have tissue-specific difference of tumourigenic effect, our discovery supports that restoring PIK3C3-mediated signaling is a rational therapeutic strategy for human CRC.
Autophagy might have dual roles in tumour progression [48]. Autophagy inhibits the migration, invasion, and metastasis of HCC and gastric cancer cells [49,50]. Beclin1 was downregulated in CRC and overexpression of Beclin1 combined with active autophagy inhibits tumour growth [51]. We found that autophagy suppresses the ras-related tumourigenesis by inhibiting cell proliferation [52] and CRC metastasis via EMT pathway. Thus, growth inhibitory effect of Rapamycin, an autophagy inducer, on CRC in vivo is rational [53]. In our prior study, rapamycin was shown to inhibit the migration of CRC cells [28]. Amiodarone, another autophagy inducer, also inhibits CRC cell migration and hepatoma cell proliferation [38]. Taken together, autophagy inducers may be considered an alternative for CRC treatment.
In conclusion, PIK3C3 mediated autophagy involves in CRC metastasis induced by miR-338-5p overexpression. The miR-338-5p/PIK3C3 ratio is a promising prognostic biomarker in the design of treatment for CRC. Further studies are needed to clarify the potential of restoring PIK3C3-mediated signaling as a therapeutic strategy for human CRC.
The following are the supplementary data related to this article.
Supplementary Fig. S2.
The effect of miR-338-5p on CRC metastasis in vivo. PIK3C3 overexpression HCT116 stable cells infected with miR-338-5p or shGFP lentivirus were injected into spleen of NOD-SCID mice and analysed for tumour metastasis. (a) Both spleen and liver were dissected out and photographed on day 42. The weight (b) and tumour volume (c) of spleen were measured. (d) Tumour cells in the ascites were examined by Liu stain. The scale bar = 200 μm. (e) miR-338-5p expression in liver nodules was assayed using QPCR. (f) Expression of PIK3C3 in primary tumours and metastasis in the liver and/or lung was examined by IHC. Data are presented as median (IQR) (n = 5) (P value was analysed using Mann-Whitney tests).
Supplementary Fig. S4.
Autophagy inhibits migration of CRC. (a) After infection with lentivirus carrying miR-338-5p or sh-GFP, HCT116 stable cells were treated with 10 μM of amiodarone for 48 h. Wound healing assays were used to analyse HCT116 stable cells counted for migrated cells after 24 h. The scale bar represents 100 μm. Data are expressed as median (IQR) (n = 6) (P value was analysed using Mann-Whitney tests). (b) SW480 cells were infected with shATG5 lentivirus. Expression of ATG5-ATG12 and LC3 was measured using Western blot. Representative results of transwell assays. The scale bar represents 50 μm. Data are presented as means ± SEM (n = 8) (P values were analysed using t tests).
Supplementary materials and methods
Supplementary tables
Acknowledgments
Acknowledgments
The authors are grateful to Dr. Wen-Chi Su for providing pCMV-VPS34 plasmids.
Funding sources
This work was supported by research grant NCKUH-10704015 from the National Cheng Kung University Hospital, Tainan, TAIWAN and grant MOST-105-2320-B-006 -029 -MY3 from the Ministry of Science and Technology, TAIWAN. The funders have no role in the study design, data collection, data analysis, interpretation, and writing of the report.
Declaration of interests
The authors declare that they have no competing financial interests.
Author contributions
C.C., C.L., H.L. and N.C. designed the experiments and wrote the paper. C.C., Y.W., C.H. and S.L. performed the experiments. C.C., C.L. and N.C. performed the immunohistochemistry experiments. C.C., J.L., Y.H., S.L., P.L., B.L. and Y.T. performed the QPCR experiment. C.C., H.L. and N.C. performed bioinformatics. All of authors have read and approved the final version of the manuscript.
Contributor Information
Hsiao-Sheng Liu, Email: a713@mail.ncku.edu.tw.
Nan-Haw Chow, Email: chownh@mail.ncku.edu.tw.
References
- 1.Global Burden of Disease Cancer C, Fitzmaurice C., Allen C., Barber R.M., Barregard L., Bhutta Z.A. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: a systematic analysis for the Global Burden of Disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markowitz S.D., Bertagnolli M.M. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeVita V.T., Hellman S., Rosenberg S.A. 7th ed. Lippincott Williams & Wilkins; Philadelphia, PA: 2005. Cancer, Principles & Practice of Oncology. [Google Scholar]
- 4.Du M., Liu S., Gu D., Wang Q., Zhu L., Kang M. Clinical potential role of circulating microRNAs in early diagnosis of colorectal cancer patients. Carcinogenesis. 2014;35:2723–2730. doi: 10.1093/carcin/bgu189. [DOI] [PubMed] [Google Scholar]
- 5.Toiyama Y., Takahashi M., Hur K., Nagasaka T., Tanaka K., Inoue Y. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen T.F., Sorensen F.B., Lindebjerg J., Jakobsen A. The predictive value of microRNA-126 in relation to first line treatment with capecitabine and oxaliplatin in patients with metastatic colorectal cancer. BMC Cancer. 2012;12(83) doi: 10.1186/1471-2407-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Igarashi H., Kurihara H., Mitsuhashi K., Ito M., Okuda H., Kanno S. Association of MicroRNA-31-5p with clinical efficacy of anti-EGFR therapy in patients with metastatic colorectal cancer. Ann Surg Oncol. 2015;22:2640–2648. doi: 10.1245/s10434-014-4264-7. [DOI] [PubMed] [Google Scholar]
- 8.Ma Y., Zhang P., Wang F., Zhang H., Yang Y., Shi C. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun. 2012;3(1291) doi: 10.1038/ncomms2276. [DOI] [PubMed] [Google Scholar]
- 9.Manceau G., Imbeaud S., Thiebaut R., Liebaert F., Fontaine K., Rousseau F. Hsa-miR-31-3p expression is linked to progression-free survival in patients with KRAS wild-type metastatic colorectal cancer treated with anti-EGFR therapy. Clin Cancer Res. 2014;20:3338–3347. doi: 10.1158/1078-0432.CCR-13-2750. [DOI] [PubMed] [Google Scholar]
- 10.Ju J.A., Huang Y.C., Lan S.H., Wang T.H., Lin P.C., Lee J.C. Identification of colorectal cancer recurrence-related microRNAs. Genomic Med Biomarkers Health Sci. 2012;4:19–20. [Google Scholar]
- 11.Kim J., Krichevsky A., Grad Y., Hayes G.D., Kosik K.S., Church G.M. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci U S A. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barik S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res. 2008;36:5232–5241. doi: 10.1093/nar/gkn513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schetter A.J., Leung S.Y., Sohn J.J., Zanetti K.A., Bowman E.D., Yanaihara N. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Huang Y., Qi Z., Sun T., Zhou Y. MiR-338-5p promotes Glioma cell invasion by regulating TSHZ3 and MMP2. Cell Mol Neurobiol. 2018;38:669–677. doi: 10.1007/s10571-017-0525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yong F.L., Law C.W., Wang C.W. Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer. 2013;13(280) doi: 10.1186/1471-2407-13-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bilegsaikhan E., Liu H.N., Shen X.Z., Liu T.T. Circulating MiR-338-5p is a potential diagnostic biomarker in colorectal cancer. J Dig Dis. 2018;19:404–410. doi: 10.1111/1751-2980.12643. [DOI] [PubMed] [Google Scholar]
- 18.Vanhaesebroeck B., Leevers S.J., Ahmadi K., Timms J., Katso R., Driscoll P.C. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 19.Lindmo K., Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- 20.Schu P.V., Takegawa K., Fry M.J., Stack J.H., Waterfield M.D., Emr S.D. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 21.Itakura E., Kishi C., Inoue K., Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lv Q., Hua F., Hu Z.W. DEDD, a novel tumor repressor, reverses epithelial-mesenchymal transition by activating selective autophagy. Autophagy. 2012;8:1675–1676. doi: 10.4161/auto.21438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lv Q., Wang W., Xue J., Hua F., Mu R., Lin H. DEDD interacts with PI3KC3 to activate autophagy and attenuate epithelial-mesenchymal transition in human breast cancer. Cancer Res. 2012;72:3238–3250. doi: 10.1158/0008-5472.CAN-11-3832. [DOI] [PubMed] [Google Scholar]
- 24.Sun Y., Liu J.H., Sui Y.X., Jin L., Yang Y., Lin S.M. Beclin1 overexpression inhibitis proliferation, invasion and migration of CaSki cervical cancer cells. Asian Pac J Cancer Prev. 2011;12:1269–1273. [PubMed] [Google Scholar]
- 25.Tuloup-Minguez V., Hamai A., Greffard A., Nicolas V., Codogno P., Botti J. Autophagy modulates cell migration and beta1 integrin membrane recycling. Cell Cycle. 2013;12:3317–3328. doi: 10.4161/cc.26298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H., Wang Y., Qian L., Wang X., Gu H., Dong X. RNF216 contributes to proliferation and migration of colorectal cancer via suppressing BECN1-dependent autophagy. Oncotarget. 2016;7:51174–51183. doi: 10.18632/oncotarget.9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu R., Yang Z., Xu G., Yu S. miR-338 modulates proliferation and autophagy by PI3K/AKT/mTOR signaling pathway in cervical cancer. Biomed Pharmacother. 2018;105:633–644. doi: 10.1016/j.biopha.2018.06.024. [DOI] [PubMed] [Google Scholar]
- 28.Ju J.A., Huang C.T., Lan S.H., Wang T.H., Lin P.C., Lee J.C. Characterization of a colorectal cancer migration and autophagy-related microRNA miR-338-5p and its target gene PIK3C3. Biomarkers Genomic Med. 2013;5:74–78. [Google Scholar]
- 29.Yeh H.H., Wu C.H., Giri R., Kato K., Kohno K., Izumi H. Oncogenic Ras-induced morphologic change is through MEK/ERK signaling pathway to downregulate Stat3 at a posttranslational level in NIH3T3 cells. Neoplasia. 2008;10:52–60. doi: 10.1593/neo.07691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeh H.H., Giri R., Chang T.Y., Chou C.Y., Su W.C., Liu H.S. Ha-ras oncogene-induced Stat3 phosphorylation enhances oncogenicity of the cell. DNA Cell Biol. 2009;28:131–139. doi: 10.1089/dna.2008.0762. [DOI] [PubMed] [Google Scholar]
- 31.Tseng Y.S., Tzeng C.C., Huang C.Y., Chen P.H., Chiu A.W., Hsu P.Y. Aurora-a overexpression associates with Ha-ras codon-12 mutation and blackfoot disease endemic area in bladder cancer. Cancer Lett. 2006;241:93–101. doi: 10.1016/j.canlet.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Besse A., Sana J., Lakomy R., Kren L., Fadrus P., Smrcka M. MiR-338-5p sensitizes glioblastoma cells to radiation through regulation of genes involved in DNA damage response. Tumour Biol. 2016;37:7719–7727. doi: 10.1007/s13277-015-4654-x. [DOI] [PubMed] [Google Scholar]
- 33.Cho D.H., Jo Y.K., Kim S.C., Park I.J., Kim J.C. Down-regulated expression of ATG5 in colorectal cancer. Anticancer Res. 2012;32:4091–4096. [PubMed] [Google Scholar]
- 34.Choi J.H., Cho Y.S., Ko Y.H., Hong S.U., Park J.H., Lee M.A. Absence of autophagy-related proteins expression is associated with poor prognosis in patients with colorectal adenocarcinoma. Gastroenterol Res Pract. 2014;2014 doi: 10.1155/2014/179586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z., Ghoorun R.A., Fan X., Wu P., Bai Y., Li J. High expression of Beclin-1 predicts favorable prognosis for patients with colorectal cancer. Clin Res Hepatol Gastroenterol. 2015;39:98–106. doi: 10.1016/j.clinre.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balgi A.D., Fonseca B.D., Donohue E., Tsang T.C., Lajoie P., Proud C.G. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PloS one. 2009;4 doi: 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lan S.H., Wu S.Y., Zuchini R., Lin X.Z., Su I.J., Tsai T.F. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014;59:505–517. doi: 10.1002/hep.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathew R., Karp C.M., Beaudoin B., Vuong N., Chen G., Chen H.Y. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hur K., Toiyama Y., Schetter A.J., Okugawa Y., Harris C.C., Boland C.R. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/dju492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong T.S., Liu X.B., Wong B.Y., Ng R.W., Yuen A.P., Wei W.I. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 42.Budhu A., Jia H.L., Forgues M., Liu C.G., Goldstein D., Lam A. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 43.Li Y., Huang Y., Qi Z., Sun T., Zhou Y. MiR-338-5p promotes glioma cell invasion by regulating TSHZ3 and MMP2. Cell Mol Neurobiol. 2018;38:669–677. doi: 10.1007/s10571-017-0525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Long J., Luo J., Yin X. MiR-338-5p promotes the growth and metastasis of malignant melanoma cells via targeting CD82. Biomed Pharmacother. 2018;102:1195–1202. doi: 10.1016/j.biopha.2018.03.075. [DOI] [PubMed] [Google Scholar]
- 45.Chen C.H., Changou C.A., Hsieh T.H., Lee Y.C., Chu C.Y., Hsu K.C. Dual inhibition of PIK3C3 and FGFR as a new therapeutic approach to treat bladder cancer. Clin Cancer Res. 2018;24:1176–1189. doi: 10.1158/1078-0432.CCR-17-2066. [DOI] [PubMed] [Google Scholar]
- 46.Chen R., Wang H., Liang B., Liu G., Tang M., Jia R. Downregulation of ASPP2 improves hepatocellular carcinoma cells survival via promoting BECN1-dependent autophagy initiation. Cell Death Dis. 2016;7:e2512. doi: 10.1038/cddis.2016.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pasquier B. SAR405, a PIK3C3/Vps34 inhibitor that prevents autophagy and synergizes with MTOR inhibition in tumor cells. Autophagy. 2015;11:725–726. doi: 10.1080/15548627.2015.1033601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su Z., Yang Z., Xu Y., Chen Y., Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer. 2015;14:48. doi: 10.1186/s12943-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weng J., Xiao J., Mi Y., Fang X., Sun Y., Li S. PCDHGA9 acts as a tumor suppressor to induce tumor cell apoptosis and autophagy and inhibit the EMT process in human gastric cancer. Cell Death Dis. 2018;9(27) doi: 10.1038/s41419-017-0189-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y., Wu P.W., Yuan X.W., Li J., Shi X.L. Interleukin-17A inhibits cell autophagy under starvation and promotes cell migration via TAB2/TAB3-p38 mitogen-activated protein kinase pathways in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2016;20:250–263. [PubMed] [Google Scholar]
- 51.Chen Z., Li Y., Zhang C., Yi H., Wu C., Wang J. Downregulation of Beclin 1 and impairment of autophagy in a small population of colorectal cancer. Dig Dis Sci. 2013;58:2887–2894. doi: 10.1007/s10620-013-2732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu S.Y., Lan S.H., Cheng D.E., Chen W.K., Shen C.H., Lee Y.R. Ras-related tumorigenesis is suppressed by BNIP3-mediated autophagy through inhibition of cell proliferation. Neoplasia. 2011;13:1171–1182. doi: 10.1593/neo.11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gulhati P., Cai Q., Li J., Liu J., Rychahou P.G., Qiu S. Targeted inhibition of mammalian target of rapamycin signaling inhibits tumorigenesis of colorectal cancer. Clin Cancer Res. 2009;15:7207–7216. doi: 10.1158/1078-0432.CCR-09-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods
Supplementary tables