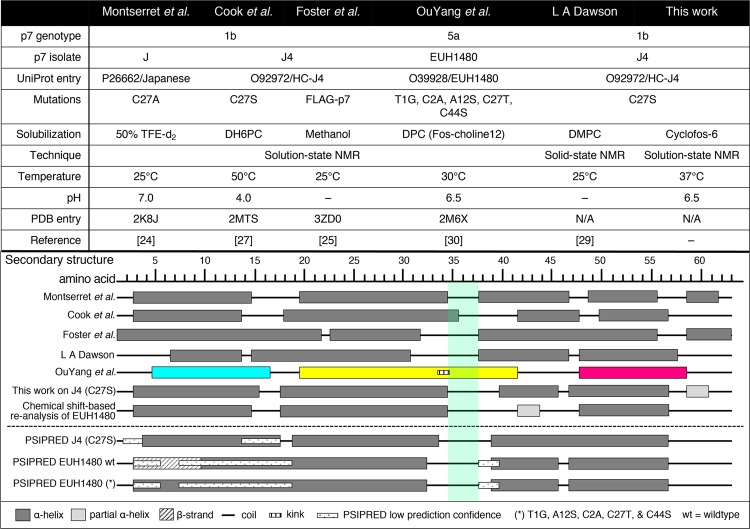
Figure 1.
Comparison of p7 secondary structures. Sample conditions (top) and secondary structure determinations (bottom) reported for p7 monomers and the hexamer of OuYang et al. The sample conditions used for p7(5a/EUH1480) in this work are the same as previously reported29. For comparison, the PSIPRED secondary structure predictions based on amino acid sequences for p7(1b/J4) and both wildtype and mutated (*) p7(5a/EUH1480) are also shown82. The PSIPRED prediction for J4 with the C27S substitution is identical to wildtype J4 (not shown). For the subunit structure reported by OuYang et al., the helices are color-coded according to the original paper29. The basic loop region ~33–37, which differs notably between monomeric genotype 1b p7 and oligomeric 5a p7, is indicated by green shading23,24,26,28,29. The secondary structures were estimated directly from published data (secondary chemical shifts, NOEs, and/or dipolar waves). In the case of p7(5a/EUH1480)29, the helical regions were determined from the structure deposited in the PDB (2M6X). Plotted helical regions by residue number: Montserret et al. 2010: 3–14, 20–34, 38–46, 48–55 and 59–61; Cook et al. 2013: 3–13, 18–35, 42–47 and 50–56; Foster et al. 2014: 1–21, 23–31, 38–55 and 59–63; LA Dawson: 7–13, 15–30, 38–46 and 48–57; Ouyang et al. 2013: 5–16, 20–41 and 48–58; Oestringer et al. 2018 J4: 3–15, 18–34, 40–45, 47–56 and 59–60; Chemical shift-based re-analysis of EUH1480 using published29 chemical shifts: 3–14, 18–34, 42–43 and 48–56; PSIPRED J4: 4–17, 19–33 and 39–56; PSIPRED EUH1480: 3–32, 39–45 and 47–56; PSIPRED EUH1480 mt5: 3–9 (beta strand), 10–32, 39–45 and 47–56.