Figure 3.
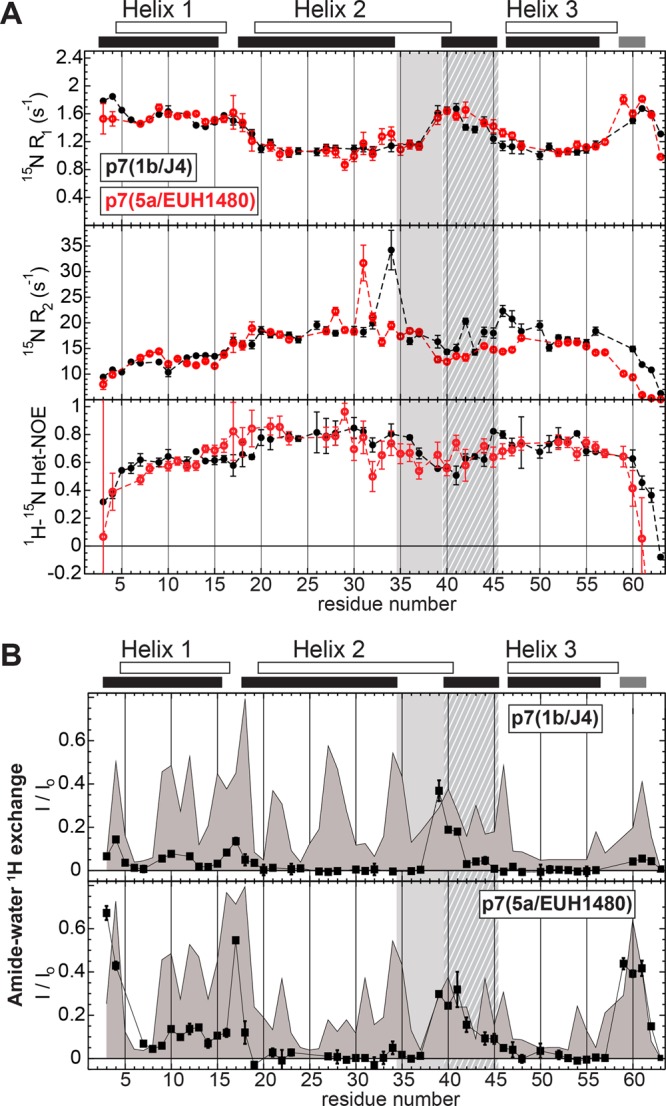
HCV p7 backbone amide dynamics and amide-water proton exchange. (A) 15N R1, 15N R2, and 1H-15N heteronuclear NOEs as a function of residue number for p7(1b/J4) (filled black circles) and p7(5a/EUH1480) (open red circles). The backbone amide heteronuclear NOE value for residue A63 of p7(5a/EUH1480) was −2.1 (data point not shown). (B) Backbone amide hydrogen exchange data for p7(1b/J4) (top) and p7(5a/EUH1480) (bottom) as a function of residue number. NMR CLEAN chemical exchange (CLEANEX) experiments83 were recorded using a 50 ms (◼) mixing time. For comparison are shown the empirically-based predictions of the magnitude of amide exchange from primary structure alone (solid line with grey shading)84. Experiments for (A,B) were recorded at 600 MHz (1H) and 37 °C. The relaxation data were collected using conventional HSQC-based pulse sequence experiments. Shown above the plots in (A,B) by unfilled and filled bars are the helical regions determined for p7(5a/EUH1480) from PDB 2M6X and those predicted from chemical shifts for p7(1b/J4), respectively. The light shading for residues 35–39 indicate helical residues in the 2M6X structural model that were found in this study to be nonhelical. The crosshatching for residues 40–45 indicate residues predicted to form an unstable helix.
