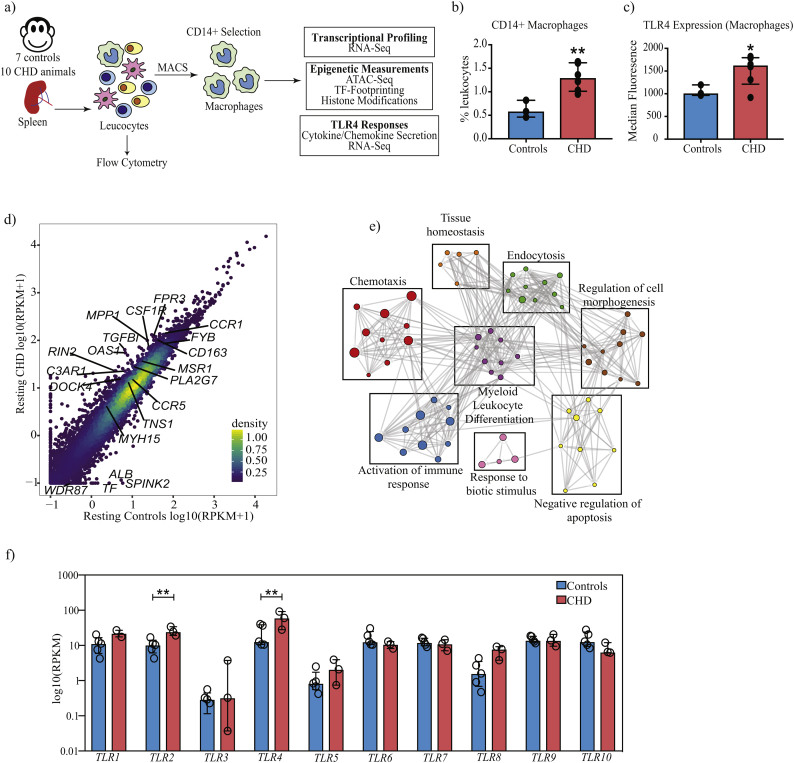
Fig. 1.
CHD is associated with transcriptional reprogramming of splenic macrophages.
(a) Schematic of the experimental design. Splenocytes from 7 control and 10 CHD animals in total were used in this study. Except for flow cytometry, all other experiments were performed on purified CD14+ splenic macrophages. (b) Bar graph depicting the percentage of CD14+ macrophages in controls (n = 3) and CHD (n = 6) animals derived from the singlet gate of splenocytes. (** - p < 0·01, unpaired t-test with Welch's correction) (c) Bar graph showing differences in TLR4 surface expression within the CD14+ splenic macrophage populations of controls (n = 3) and CHD (n = 6) animals measured using flow cytometry. Median Fluorescence Intensity (MFI) is depicted on the Y-axis. (* - p < 0·05, unpaired t-test with Welch's correction) (d) Scatterplot comparing average normalised transcript counts (RPKM) from resting splenic macrophages in response to CHD. Select significantly differentially expressed genes (DEG) outside the confidence interval of the linear fit are highlighted. A total of 5 controls and 3 CHD animals were used in the RNA-Seq experiment. (e) Functional enrichment of DEG detected in resting splenic macrophages in response to CHD was carried out using Metascape. Each circle denotes a Gene Ontology (GO) term. The size of the circle indicates the number of DEG enriching to the GO term in question. Closely related (GO) terms are clustered into groups labelled with the most statistically significant GO term within that group. Grey lines indicate the relationship between different GO terms based on shared DEG. (f) Bar graphs showing normalised transcript counts (RPKM) of Toll-like receptors genes (TLR) (** - FDR <0·01, exact test in edgeR followed by FDR correction using Benjamini-Hochberg method).