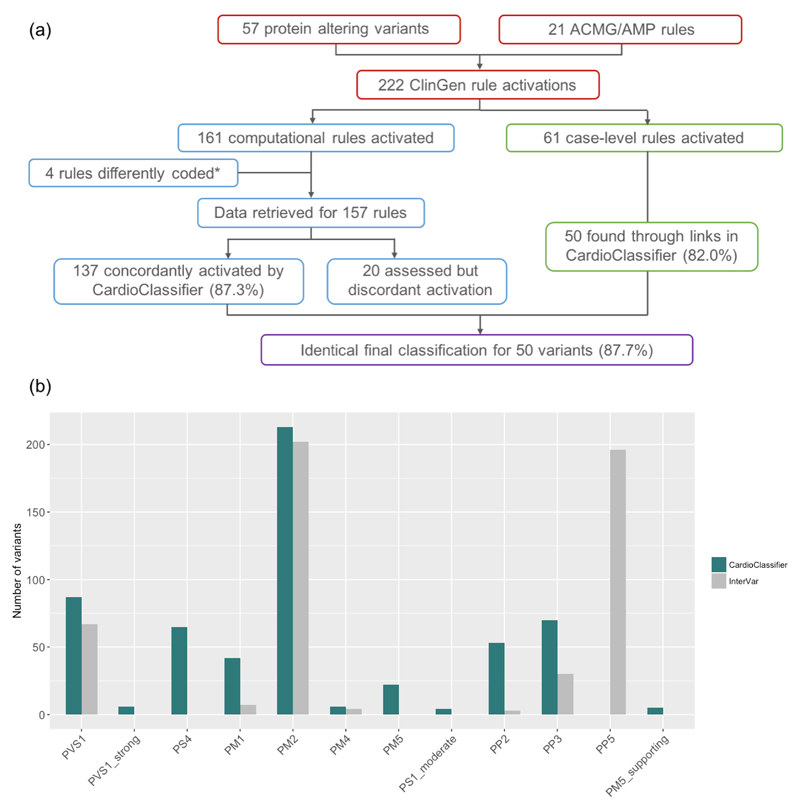
Figure 3. Validation of CardioClassifier.
(a) Comparing CardioClassifier to a set of 57 MYH7 expert panel curated variants. Rules were split into those that can be computationally annotated and those that are 'case-level' and require manual input. CardioClassiifer was run using an 'All Cardiomyopathy' test to reflect the spectrum of phenotypes caused by variants in MYH7. *Of the computational rules, 3 were removed from the comparison as they represent draft modifications to the ACMG framework by the ClinGen Cardiovascular domain working group that were not published at the time of this work, and not yet implemented in CardioClassifier. Specifically, truncating variants in MYH7 activate a new rule PVS1_moderate. Additionally, for variants classified as Benign by frequency alone (BA1) CardioClassifier does not assess any further rules, leading us to remove an additional data point from the comparison as we would not expect it to be retrieved. (b) Counts of individual rules activated by CardioClassifier and InterVar for 219 variants identified as Pathogenic or Likely Pathogenic in ClinVar. Only pathogenic evidence rules and rules activated by one of the tools at least once are shown.