Figure 1: Recovering thousands of de novo engineered L1 retrotransposition events.
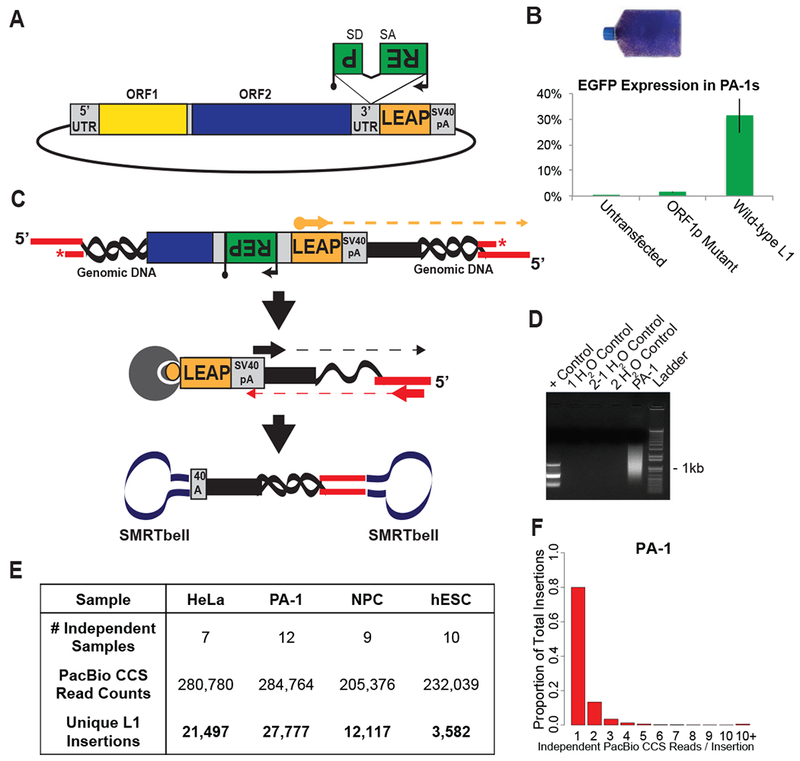
(A) Engineered human L1 expression plasmids contain a retrotransposition indicator cassette (mneoI or mEGFPI) within their 3’UTR (green rectangle with the backward ‘REP’ for ‘Reporter’). The reporter (black arrow, promoter; black lollipop, polyadenylation signal) is in the opposite transcriptional orientation of the L1 and is interrupted by an intron (SD, splice donor; SA, splice acceptor) in the same transcriptional orientation as the L1.
(B) Representative flask of G418-resistant HeLa-JVM cells (top), and the proportion of FACS-sorted EGFP-positive PA-1 cells. Untransfected cells and an L1 ORF1p mutant served as negative controls.
(C) Genomic DNA isolated from cells harboring L1 integration events was sheared and ligated to adapters containing a blocking 3’ amine group (red asterisk). Linear amplification utilized a biotinylated primer specific to the engineered L1 (orange arrow). Products were captured on streptavidin beads (gray circle) and subjected to nested PCR utilizing primers specific to the SV40pA signal (black arrow) and ligated adapter (red arrow). Ligation of SMRTbell adapters (navy dumbbells) facilitated PacBio CCS sequencing.
(D) Gel image of a library created from PA-1 cells shows a smear indicative of many recovered L1 insertions (Lane 5). Lane 1, expected products for a parallel PC39 positive control preparation. Lanes 2 to 4 are water blanks.
(E) Numbers of independent samples, PacBio CCS reads, and unique L1 insertions obtained from the four analyzed cell lines.
(F) Frequency distribution of the number of independent CCS reads (i.e., those with different shear points) supporting L1 insertion events from PA-1 cells.
