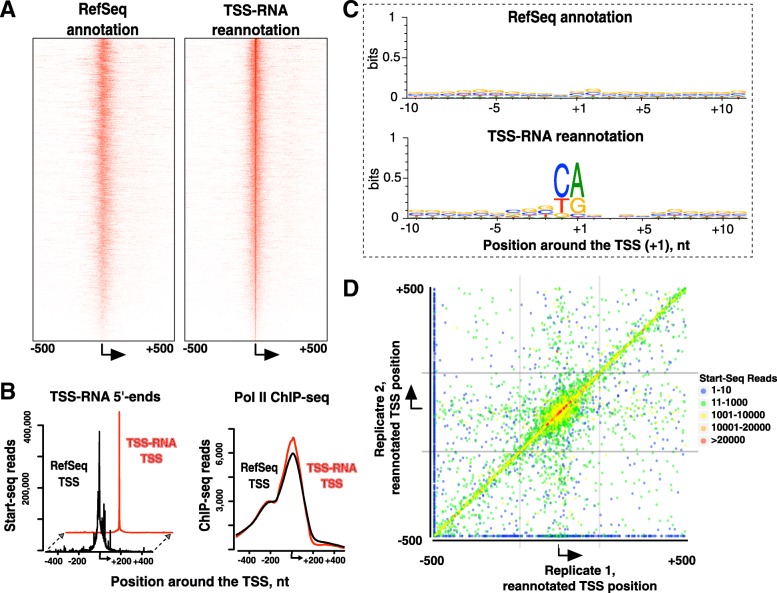
Fig. 2.
TSS RNA-based refinement of gene TSSs. a. Heatmaps of sense-strand TSS-RNA on 7112 rat genes ordered by decreasing TSS-RNA count within the promoter region, centered around annotated (left panel) and TSS-RNA peak-centered locations (right panel). b. Left pane. Metaplots of TSS-RNA 5′-ends for the same genes as in A, centered on TSSs defined using RefSeq (black) and TSS-RNA (red) TSS annotations. Right pane. Pol II ChIP-sequencing traces based on previously published data [10] centered against the same TSSs. c. Weblogo visualization of sequence enrichment of the genes centered around RefSeq annotation (top) and TSS-RNA reannotation (bottom panel). d. Correspondence between TSS annotations between two independent biological replicates, with each dot representing a gene and color coding indicating the number of TSS-RNA hits per gene as indicated. Genes are plotted against RefSeq-annotated TSSs. Strikethrough lines enclose genes considered for TSS reannotations based on each replicate, with the center quadrant containing genes that were reannotated