Abstract
Background
Methicillin-resistant Staphylococcus aureus (MRSA) is known as a leading cause of morbidity and mortality. Investigation of the MRSA’s virulence and resistance mechanisms is a continuing concern toward controlling such burdens through using high throughput whole Genome Sequencing (WGS) and molecular diagnostic assays. The objective of the present study is to perform whole-genome sequencing of MRSA isolated from Sudan using Illumina Next Generation Sequencing (NGS) platform.
Results
The genome of MRSA strain SO-1977 consists of 2,827,644 bp with 32.8% G + C, 59 RNAs and 2629 predicted coding sequences (CDSs). The genome has 26 systems, one of which is the major class in the disease virulence and defence. A total of 83 genes were annotated to virulence disease and defence category some of these genes coding as functional proteins. Based on genome analysis, it is speculated that the SO-1977 strain has resistant genes to Teicoplanin, Fluoroquinolones, Quinolone, Cephamycins, Tetracycline, Acriflavin and Carbapenems. The results revealed that the SO-1977, strain isolated from Sudan has a wide range of antibiotic resistance compared to related strains.
Conclusion
The study reports for the first time the whole genome sequence of Sudan MRSA isolates. The release of the genome sequence of the strain SO-1977 will avail MRSA in public databases for further investigations on the evolution of resistant mechanism and dissemination of the -resistant genes of MRSA.
Keywords: Methicillin -resistance Staphylococcus aureus (MRSA), Whole genome sequencing, Antibiotic resistant genes, Genome annotation, Sudan
Background
Staphylococcus aureus (S. aureus) is a human pathogen known to cause both nosocomial and community-acquired infections [1]. It has been identified, among other classes of bacteria, resistant against some antibiotics. One of the emerged resistant strain of S. aureus is Methicillin-resistant Staphylococcus aureus (MRSA) that is the leading cause of life-threatening infections even in countries with advanced health surveillance and maintenance systems [1, 2]. In Sudan, MRSA’s incidence rate has increased dramatically and has been reported to be associated with wound infection constituting substantial sources of the high morbidity and mortality rate [3]. Such emergence of resistant strains is due to the overuse of not developed antibiotics that ultimately makes real challenges at treatment. Therefore, there is an urgent need to uncover the genetic basis of their virulence and resistance mechanism for better understanding as well as addressing potential effective drug targets. Over the last decades, Whole-genome sequencing (WGS) technologies witnessed large volumes of produced data including mutant genes, cancer-causing genes and genes predisposing for certain diseases. Moreover, the advanced bench-top sequencers technique, applied in regular clinical laboratories [4] may result in enormous diagnostic developments and challenges [5]. Genomic materials of S. aureus strains have been studied to understand the mechanisms and virulence factors responsible for staphylococcal antibiotic resistance. The premier S. aureus genomes sequenced were; MRSA strains N315 and Mu50 [6] followed by other nine strains [7, 8]. The studies revealed that the length of staphylococcal genomes is about 2.8 Mbp with low GC content. The regions of staphylococcal genomes are well conserved, with many massive sequence blocks showing high variability [8]. Although a considerable number of the MRSA resistant to antimicrobials including Methicillin, Ofloxacin, Penicillin, Amikacin, and Vancomycin are reported in Sudan [9], the molecular investigations that help in understanding the mechanism of MRSA epidemics at the whole genome level are yet limited. The present study aims to analyse the whole genome sequence (WGS) of SO-1977 strain and subsequently evaluates the genomic diversity and genotypic prediction of the antimicrobial resistance of MRSA isolated from a patient in Sudan.
Results
Genome project history
The genome sequences of SO-1977 strain were deposited in GenBank® (WGS database). The result was summarized in (Table 1).
Table 1.
Project Information
| Property | SO-1977 |
|---|---|
| Finishing quality | Complete |
| Libraries used | 2 × 250 bp |
| Sequencing platforms | Illumina MiSeq |
| Fold coverage | 122.26x |
| Assembly Method | SPAdes v. 3.9.0 |
| GenBank ID | NFZY00000000 |
| GenBank date of Release | 27-JUL-2017 |
| BIOPROJECT | PRJNA385553 |
| BioSample | SAMN06894057 |
| Locus Tag | CA803 |
| Source Material Identifier | Wound |
| Project relevance | Medical |
Genomic features of strain SO-1977
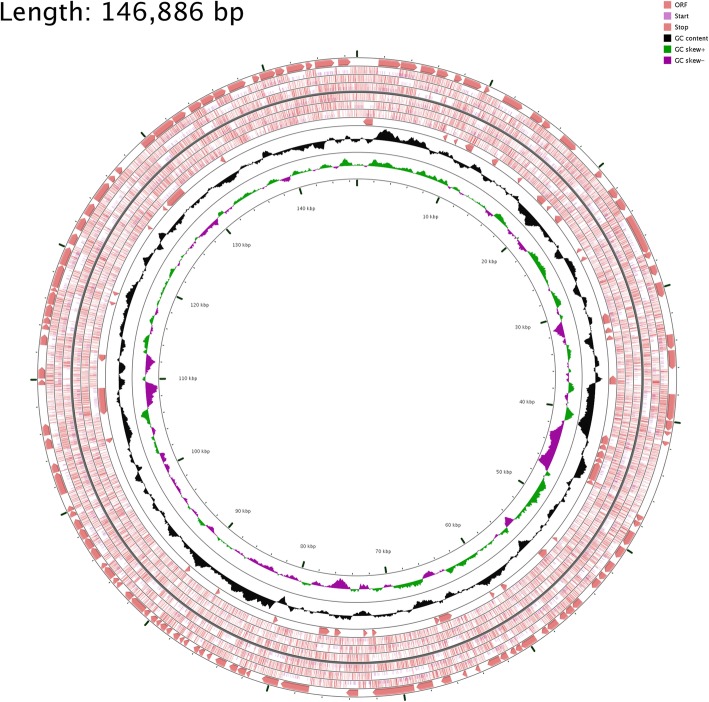
As can seem from the data in Table 2, the draft genome sequence of S. aureus strain SO-1977 consisted of 2,827,644 bp with a 32.8% GC. The number of predicted coding sequences (CDS), tRNAs and rRNAs was 2629, 51 and 4 respectively. The final assembly contained 151 contigs with N50 of 62,783 bp length. The largest contig assembled was 146,886 bp length.
Table 2.
Nucleotide and gene content’s levels of the MRSA SO-1977 genome
| Attribute | Value |
|---|---|
| Genome size (bp) | 2,827,644 bp |
| DNA G + C content | 32.8% |
| Number of Contigs | 151 |
| N50 | 62,783 bp |
| rRNA genes | 4 |
| tRNA genes | 51 |
| ncRNAs | 4 |
| Protein-coding genes | 2629 |
| Pseudo Genes (total) | 129 |
| Pseudo Genes (frameshifted) | 62 |
| Pseudo Genes (incomplete) | 32 |
| Pseudo Genes (internal stop) | 50 |
| Pseudo Genes (multiple problems) | 13 |
| Genes assigned to SEED | 1698 |
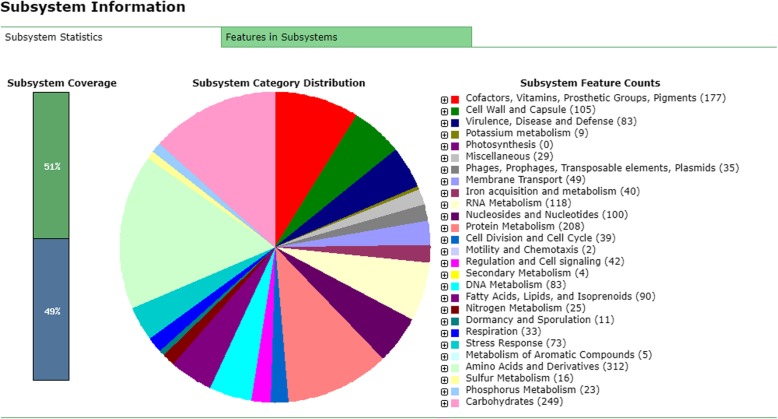
Genome annotation using RAST (Fig. 1)
Fig. 1.
Summary of annotation for MRSA strain SO-1977 based on RAST subsystem
Whole-genome annotation of MRSA strain SO-1977 on RAST server revealed a total of 1970 genes belonging to 26 subsystems such as Cofactors, Vitamins, Prosthetic Groups, Pigments, Cell Wall and Capsule and Virulence, Disease and Defense. The graphical circular map of the SO-1977 genomes was shown in Fig. 2.
Fig. 2.
Circular map of the chromosome of the S. aureus SO − 1977. The innermost ring represents the SO − 1977 chromosome. The second ring (in black) plots the G + C content of the reference, followed by its G + C skew (in purple/green)
Genes involved in virulence, disease and defence
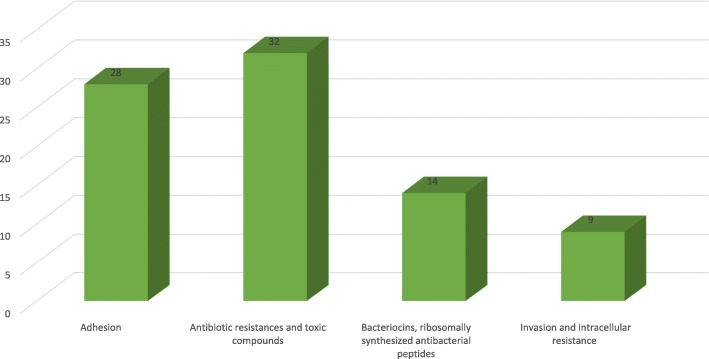
Result revealed that 83 genes encoded for virulence, disease, and defence, 28 genes were annotated to be responsible for adhesion, 32 for antibiotic resistances and toxic compounds, 14 for Bacteriocins, ribosomally synthesized antibacterial peptides and 9 for invasion and intracellular resistance (Fig. 3). Some of these genes which coding functional proteins are Fibronectin binding protein, Chaperonin, Two-component response regulator BceR, Folylpolyglutamate synthase, Acetyl-coenzyme A, Carboxyl transferase beta chain, Colicin V production protein, MerR family, Multidrug resistance protein, Mercuric ion reductase and Arsenate reductase. The category of the cell wall and capsule system of peptidoglycan biosynthesis revealed that two genes have a relationship with conferring Methicillin resistance while one gene was related to Penicillin resistance.
Fig. 3.
Bar diagram. Genes involved in category virulence disease and defense
Phages, prophages, transposable elements, plasmids (Table 3)
Table 3.
Systems included Phages, prophages, transposable elements, plasmids category
| Category | Subcategory | Subsystem | Role |
|---|---|---|---|
| Phages, Prophages, Transposable elements, Plasmids | Phages, Prophages | Phage tail proteins | Phage tail protein/ Phage tail length tape-measure protein |
| Phage replication | Single stranded DNA-binding protein/ Phage replication initiation protein/ DNA polymerase III alpha subunit/ DNA helicase, phage-associated | ||
| Phage packaging machinery | Phage DNA packaging/ Phage terminase, small subunit/ Phage terminase, large subunit/ Phage portal protein | ||
| Phage capsid proteins | Phage capsid and scaffold/ Phage head maturation protease/ Phage major capsid protein/ Phage capsid protein | ||
| Phage lysis modules | Phage lysin, N-acetylmuramoyl-L-alanine amidase/ Phage holin | ||
| Pathogenicity islands |
Listeria Pathogenicity Island LIPI-1 extended |
Phosphatidylinositol-specific phospholipase | |
| Zinc metalloproteinase precursor |
The analysis revealed that 35 genes are encoding for Phages, Prophages, Transposable elements, Plasmid of which 33 were annotated to be responsible for Phages, Prophages and Pathogenicity islands.
Resistant genes based comparative genomic analysis (Table 4)
Table 5.
List of 16S rRNA
| Strain | Accession no. | Seq match |
|---|---|---|
| S. aureus | L37597.1 | 1 |
| S. aureus MPU99 | AB353073.1 | 1 |
| S. aureus SA1 | AB305019.1 | 1 |
| S. aureus ATCC 14458 | DQ997837.1 | 1 |
| S. aureus subsp. anaerobius DSM 20714 | AY688031.1 | 1 |
| S. aureus K16–13 | AY859409.1 | 1 |
| S. aureus1 | AY144447.1 | 1 |
aSeq match score, the number of (unique) 7-base oligomers shared between your sequence and a given RDP sequence divided by the lowest number of unique oligos in either of the two sequences
The Genome annotation and comparison results by RSAT server have shown that SO-1977 strain possesses 29 genes that may be related to multi-drug resistance and the comparison between MRSA strains was shown that 23 resistant genes were present in all strains, two genes were only found in SO-1977 strain conferring resistance against Tetracycline. Furthermore, The SO-1977 strain was the only one having the norA gene providing resistance against Quinolone beside other six genes of the family MarR. Four genes that are responsible for anti- Methicillin resistance (LytH, MecI, Mec and MurE) were only found in MRSA252 strain. Also the results have shown that MRSA252 and MSSA476 are sharing a single common gene for anti-Methicillin resistance (HmrB).
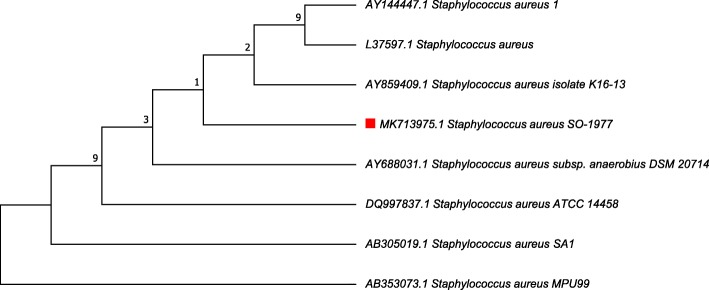
Phylogenetic analysis of nucleotide sequence of strain SO-1977
Result on the phylogenetic of 16S rRNA (MK713975) showed that the SO-1977 strain has the highest similarity with different S. aureus strain (Table 5) (Fig. 4).
Table 4.
Summary of CDSs annotated to antibiotic resistance between SO-1977, MRSA252 and MSSA476 strains
| No | SO-1977 | MRSA252 | MSSA476 | Seed subsystem | Seed function | Length (bp) | Contig number |
|---|---|---|---|---|---|---|---|
| 1 | ✓ | ✓ | Peptidoglycan Biosynthesis | Methicillin resistance determinant MecA, transpeptidase | 2007 | Contig 000034 | |
| 2 | ✓ | ✓ | ✓ | Multidrug Resistance, 2-protein version Found in Gram-positive bacteria | Multidrug resistance protein [function not yet clear] | 648 | Contig 000037 |
| 3 | ✓ | ✓ | ✓ | Multidrug Resistance, 2-protein version Found in Gram-positive bacteria | Membrane component of multidrug resistance system | 1932 | Contig 000037 |
| 4 | ✓ | ✓ | ✓ | Multidrug Resistance, 2-protein version Found in Gram-positive bacteria | TetR family regulatory protein of MDR cluster | 555 | Contig 000037 |
| 5 | ✓ | Tetracycline resistance, ribosome protection type | Tetracycline resistance protein TetM | 1920 | Contig 000042 | ||
| 6 | ✓ | Tetracycline resistance, ribosome protection type | Translation elongation factor G | 2082 | Contig 000014 | ||
| 7 | ✓ | ✓ | ✓ | Teicoplanin-resistance in Staphylococcus | Teicoplanin resistance associated membrane protein TcaA | 1209 | Contig 000002 |
| 8 | ✓ | ✓ | ✓ | Teicoplanin-resistance in Staphylococcus | Teicoplanin resistance associated membrane protein TcaB | 1383 | Contig 000002 |
| 9 | ✓ | ✓ | ✓ | Teicoplanin-resistance in Staphylococcus | Teicoplanin-resistance associated HTH-type transcriptional regulator TcaR | 456 | Contig 000002 |
| 10 | ✓ | Quinolone resistance protein norA | none | 1167 | Contig 000007 | ||
| 11 | ✓ | ✓ | Bicyclomycin resistance protein TcaB | none | 1212 | Contig 000002 | |
| 12 | ✓ | ✓ | ✓ | Transcriptional regulator, MarR family | None | 456 | Contig 000003 |
| 13 | ✓ | ✓ | ✓ | Transcriptional regulator, MarR family | None | 420 | Contig 000005 |
| 14 | ✓ | ✓ | ✓ | Transcriptional regulator, MarR family | None | 210 | Contig 000043 |
| 15 | ✓ | ✓ | ✓ | Transcriptional regulator, MarR family | None | 435 | Contig 000030 |
| 16 | ✓ | ✓ | ✓ | Transcriptional regulator, MarR family | None | 441 | Contig 000001 |
| 17 | ✓ | ✓ | ✓ | Transcriptional regulator,MarR family | None | 468 | Contig 000030 |
| 18 | ✓ | ✓ | ✓ | Resistance to fluoroquinolones | DNA gyrase subunit B | 2403 | Contig 000010 |
| 19 | ✓ | ✓ | ✓ | Resistance to fluoroquinolones | DNA gyrase subunit A | 2661 | Contig 000011 |
| 20 | ✓ | ✓ | ✓ | Resistance to fluoroquinolones | Topoisomerase IV subunit B | 1998 | Contig 000010 |
| 21 | ✓ | ✓ | ✓ | Resistance to fluoroquinolones | Topoisomerase IV subunit A | 2403 | Contig 000010 |
| 22 | ✓ | ✓ | ✓ | Beta-lactamase | Beta-lactamase | 1500 | Contig 000002 |
| 23 | ✓ | ✓ | ✓ | Beta-lactamase | Beta-lactamase repressor BlaI | 381 | Contig 000067 |
| 24 | ✓ | ✓ | ✓ | Multidrug Resistance Efflux Pumps | Acriflavin resistance protein | 3168 | Contig 000001 |
| 25 | ✓ | ✓ | ✓ | Multi antimicrobial extrusion protein (Na(+)/drug antiporter), MATE family of MDR efflux pumps | Multidrug Resistance Efflux Pumps | 1356 | Contig 000005 |
| 26 | ✓ | ✓ | Methicillin resistance in staphylococci | HmrB protein involved in methicillin resistance | 1600 | NC_002952 | |
| 27 | ✓ | ✓ | ✓ | Methicillin resistance in staphylococci | Beta-lactamase repressor BlaI | 381 | Contig 000067 |
| 28 | ✓ | ✓ | ✓ | Methicillin resistance in staphylococci | FmhA protein of FemAB family | 1251 | Contig 000002 |
| 29 | ✓ | Methicillin resistance in staphylococci | LytH protein involved in methicillin resistance | 1600 | NC_002952 | ||
| 30 | ✓ | ✓ | Methicillin resistance in staphylococci | Methicillin resistance regulatory sensor-transducer MecR1 | 1600 | Contig 000034/ NC_002952 | |
| 31 | ✓ | Methicillin resistance in staphylococci | Methicillin resistance repressor MecI | 1600 | NC_002952 | ||
| 32 | ✓ | Methicillin resistance in staphylococci | Transposase for insertion sequence-like element IS431mec | 1296 | NC_002952 | ||
| 33 | ✓ | Methicillin resistance in staphylococci | UDP-N-acetylmuramoylalanyl-D-glutamate--2,6-diaminopimelate ligase | 1600 | NC_002952 | ||
| 34 | ✓ | ✓ | ✓ | Methicillin resistance in staphylococci | Beta-lactamase regulatory sensor-transducer BlaR1 | 1089 | Contig 000067 |
Fig. 4.
Neighbour-joining tree based on 16S rRNA gene sequences showing the Phylogenetic relationship between Staphylococcus aureus strain SO-1977 relative to other type strains within the Staphylococcus aureus in database. Bootstrap values (expressed as percentages of 100 replications) less than 50% are hidden
Discussion
The present study reported the first genome sequence of S. aureus (MRSA) isolated from Sudan to have phylogenetic allocation using the 16S rRNA gene to represent the evolutionary relationships of the bacteria. In this study, the phylogenetic analysis of the complete 16S rRNA gene sequence of strain SO-1977 (MK713975) has shown that the strain should be assigned to the genus Staphylococcus. The annotated draft genome sequence of SO-1977 strain was 2827,644 bp length containing 2629 coding sequences (CDS). Moreover, the WGS data was used to investigate antimicrobial resistance and virulence mechanism. The multi-drug resistance of this isolate might be generated by the ability of these bacteria to accumulate multiple genes on the resistance (R) plasmids coding for a single drug resistance within a single cell or by the increased expression of genes that code for multi-drug efflux pumps, extruding a wide range of drugs [10]. In this study, S. aureus (MRSA) isolated from Sudan has been demonstrated to possess different resistance mechanisms which can be attributed to the use of resistant genes TcaR, TcaA, TcaB, TetR, TetM, PBP2a (MecA), or by secretion of enzymes (DNA gyrase subunit A, DNA gyrase subunit B, Topoisomerase IV subunit A, Topoisomerase IV subunit B and Beta-lactamase repressor) allowing it to use the efflux pump mechanism. In addition, six putative MarR family transcriptional regulators in the SO-1977 genome were identified. These were recognised as a widely conserved group of multiple antibiotic resistance regulators that respond to a wide range of antibiotics [11]. The MRSA characteristic phenotype is due to the presence of mecA, which encodes a penicillin-binding protein (PBP), PBP2a, with reduced affinity for b-lactams. MecA is embedded in a large heterologous chromosomal cassette, the SCCmec element. Some MRSA strains carry upstream to the mecA gene such as the regulatory genes mecI-mecR1 that encoding for a repressor and a sensor/inducer of the mecA expression, respectively [12]. In this study, MecA and MecR1 were found in SO-1977 and MRSA252, while the mecI was found only in MRSA252. This result suggested that the existence of yet unidentified additional determinants involved in the transcriptional control of mecA gene and point to a revision of the mecA regulatory mechanism in MRSA SO-1977 strain. The antibiotic sensitivity tests demonstrated that the isolate is resistant to discsoxacillin and cefoxitin. The result of the WGS confirmed the resistance of the isolate to the antibiotics and expanded it to include Teicoplanin, Fluoroquinolones, Quinolone, Cephamycins, Tetracycline, Acriflavin and Carbapenems. Such a result should be considered while planning an effective treatment protocol. The antibiotic-resistant genes of SO-1977, MRSA252 and MSSA476 revealed that the SO-1977 strain isolated from Sudan is complicated and has a wide range of cross-antibiotic resistance.
Conclusion
The current study is the first of its kind in Sudan to give an insight of an important antibiotic resistant bacterial strain, MRSA SO-1977. The SO-1977 strain is resistant to Teicoplanin, Fluoroquinolones, Quinolone, Cephamycins, Tetracycline, Acriflavin and Carbapenems. This study strongly suggests that other yet unidentified determinants are involved directly or indirectly in the transcriptional control of the mecA gene in SO-1977 strain. The SO-1977 strain has a wide range of antibiotic resistance compared to other strains. The whole genome of SO-1977 strain can provide a genetic background of virulence, antibiotic resistance and Phages of the MRSA species in Sudan.
Methods
Sample preparation
A wound swab specimen was collected from a patient at Soba Hospital, Khartoum, and was inoculated in sheep blood agar and mannitol salt agar at 37 °C for 24 h. For the purpose of colonies identification, standard procedures and tests were performed including gram stain, catalase, coagulase, and DNase tests were used to identify the colonies [13]. The positive cultures for S. aureus were then suspended with a concentration similar to turbidity standard equivalent to 0.5% McFarland and streaked on Mueller-Hinton agar (MHA). Oxacillin (6μg\ml) and cefoxitin (30μg\ml) antimicrobial disc were positioned at suitable distances on the bacterial lawns on MHA at 33 °C for 24 h. The antibiotic resistance profiling of the strain against a broader range of antibiotics was not performed as a limitation of the study. The growth inhibition zones were then measured according to the standard Kirby –Bauer disc diffusion method and NCCLs guidelines using a calliper [14]. In which the revealed measurements were indicatives of resistant colonies of MRSA strain.
Genomic DNA extraction and sequencing library preparation
Bacterial DNA was extracted using Qiagen Kit following the manufacturer instructions. The concentration and purity of the resultant DNA were photo-metrically determined using a Nano-drop (Thermofisher®). About 5 μg of genomic DNA (A260/280 = 1.93) was used for library preparation and 4 nm of genomic DNA was used as an input for the Nextera XT kit (Illumina). Then samples were targeted for bar-coding using forward (N702) and reverse (N702) primers in 12 cycles of amplification in the PCR machine. Libraries were then quantified on the Bioanalyzer (Agilent Technologies) and combined with an equimolar mixture. Finally, 0.19 ng/ ml was used as an input for Next-generation sequencing (NGS) and libraries were sequenced on a single run on the Illumina MiSeq instrument (250 bp paired-end reads).
Bacterial genome sequencing and assembly
Poor-quality and adaptor-containing reads were filtered and trimmed using BBTools version 36 [15]. Good quality sequencing reads were assembled using SPAdes version 3.5.0. For the prediction of tRNA and rRNA genes, ARAGORN 1.2.34 and RNAmmer1.2 were used, respectively [16, 17]. The protein-coding genes were then predicted using Prodigal 2.60 [18] as well as their function by using BLASTN 2.2.25+ [19] and followed by detecting sequence homologs through searching for various sequence domain databases using HMMER 3.0 (http://hmmer.org/).
Genome annotation
The final draft genome sequence of S. aureus SO-1977 was used for annotation using RAST [20] and NCBI Prokaryotic Genome Annotation Pipeline [21]. The annotated genes were exported from the RAST server into an excel table and manually compared for genomic features. The antibiotic resistance genes of the S. aureus SO-1977, S. aureus MRSA252 (PRJNA265) and S. aureus MSSA476 (PRJNA116329) were retrieved from RAST server then the comparison was done [22]. The graphical circular map of the genomes was made by CGView server [23].
Phylogenetic analysis of strain SO-1977 using 16S rRNA
The 16S rRNA sequences were edited and assembled to a final length of 1545 bp, GenBank accession number (MK713975). All reference sequences of the 16S rRNA genes of S.aureus used in this study were obtained from GenBank® (https://www.ncbi.nlm.nih.gov/genbank/) using the RDP 11.5, Seqmatch: version 3 (https://rdp.cme.msu.edu/). DNA sequence alignment was performed using MUSCLE v. 3.8.31 (http://www.ebi.ac.uk/Tools/msa/muscle/) on the European Bioinformatics Institute (EBI) homepage. Once the alignment was completed, the phylogenetic tree was drowning as the evolutionary distances were computed using the Maximum Likelihood method implemented in MEGA6 version 6 [24] in all positions containing gaps and missing data were eliminated.
Sequence data access
The genomic data of this study were deposited publicly in DDBJ/ENA/GenBank® under Accession: NFZY00000000, BioProject: PRJNA385553 and Biosample: SAMN 06894057.
Acknowledgements
We thank Prof. Mohamed M.A Alnour and Ms. Somyia Kambal for reviewed the manuscript. This work was supported by National University Research Institute-National University, Sudan.
Funding
This work was supported by National university research institute / National University. The funding body had a role.
Availability of data and materials
The datasets generated during the current study are available from the corresponding author publicly available due (Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession NFZY00000000 and PRJNA385553. The version described in this paper is version NFZY01000000.
Abbreviations
- CDS
Coding sequences
- MRSA
Multidrug-resistant Staphylococcus aureus
- NGS
Next-generation sequencing
- WGS
Whole genome sequencing
- RAST
Rapid Annotation using Subsystem Technology
Authors’ contributions
SBM, MSA and AOS planned and directed the project. SEM and MIG collected the sample, TBA, AEA, ZSAA and FMA laboratory work and bacterial identification. SBM, RAO and NMI drafted the manuscript. SBM, N-SL and AAA performed the gene annotation and comparative genomic analysis and interpreted the results. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This research was approved by the Ethical Committee of the International University of Africa, Khartoum Sudan and written consent was obtained.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohamed S. Ali, Email: alkhatimali@gmail.com
Nurulfiza M. Isa, Email: nurulfiza@upm.my
Faisal M. Abedelrhman, Email: faisalmousa86@gmail.com
Tahani B. Alyas, Email: tonaarabi4@yahoo.com
Sara E. Mohammed, Email: srora48@gmail.com
Abdallah E. Ahmed, Email: abdallah_elseer@yahoo.com
Zainab S. A. Ahmed, Email: zasaali9@gmail.com
Nyok-Sean Lau, Email: nyoksean@usm.my.
Mohamed I. Garbi, Email: mogh511@gmail.com
Abdullah Al-Ashraf Amirul, Email: amirul@usm.my.
Almeen O. Seed, Email: aminsaba@hotmail.com
Rihab A. Omer, Email: rihab.omer@yahoo.com
Sofia B. Mohamed, Phone: +249966686656, Email: Sofiabashir2002@gmail.com
References
- 1.Naimi Timothy S., LeDell Kathleen H., Boxrud David J., Groom Amy V., Steward Christine D., Johnson Susan K., Besser John M., O’Boyle Carol, Danila Richard N., Cheek James E., Osterholm Michael T., Moore Kristine A., Smith Kirk E. Epidemiology and Clonality of Community‐Acquired Methicillin‐ResistantStaphylococcus aureusin Minnesota, 1996–1998. Clinical Infectious Diseases. 2001;33(7):990–996. doi: 10.1086/322693. [DOI] [PubMed] [Google Scholar]
- 2.Torell Erik, Molin Daniel, Tano Eva, Ehrenborg Christian, Ryden Cecilia. Community-acquired pneumonia and bacteraemia in a healthy young woman caused by methicillin-resistant Staphylococcus aureus (MRSA) carrying the genes encoding Panton-Valentine leukocidin (PVL) Scandinavian Journal of Infectious Diseases. 2005;37(11-12):902–904. doi: 10.1080/00365540500348945. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed RA. Detection of methicillin-resistant Staphylococcus aureus associated with wound infections in Khartoum teaching hospital, Sudan. Afr J Med Sci. 2016;1(11 suppl):1–4.
- 4.Didelot X, Bowden R, Wilson DJ, Peto TE, Crook DW. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet. 2012;13:601–612. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metzker ML. Sequencing technologies - the next generation. NatRev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 6.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J-Q, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Kobayashi K, Tanaka T, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa K, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. Whole genome sequencing of methicillin resistant Staphylococcus aureus. Lancet. 2001;357(9264):1225–1240. doi: 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 7.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan MRJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. Complete genome sequence of USA300, an epidemic clone of acquired methicillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 9.Kheder SI, Ali NA, Fathelrahman AI. Prevalence and Antimicrobial Susceptibility Pattern of Methicillin Resistance Staphylococcus in a Sudanese Surgical Ward. Pharmacol Pharm. 2012;3:103–108. doi: 10.4236/pp.2012.31015. [DOI] [Google Scholar]
- 10.Chambers HF, DeLeo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7(9):629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Lei M, Song Y, Gong K, Li L, Liang H, Jian X. Whole genome sequence and comparative genomic analysis of multidrug-resistant Staphylococcus capitis subsp. urealyticus strain LNZR-1. Gut Pathog. 2014;6:45. doi: 10.1186/s13099-014-0045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliveira DC, de Lencastre H. Methicillin-resistance in Staphylococcus aureus is not affected by the overexpression in trans of the mecA gene repressor: a surprising observation. PLoS One. 2011;6(8):e23287. doi: 10.1371/journal.pone.0023287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergy, Divid H, Holt JG, Krieg NR, Peter HA. Sneath, Bergy's manual of determinative bacteriology. 9. Phladelphia: Lippincott Williams and Wilikins; 1994. [Google Scholar]
- 14.Wayne PA. Clinical and laboratory standards institute/NCCL. 2006. [Google Scholar]
- 15.Bartels M. D., Petersen A., Worning P., Nielsen J. B., Larner-Svensson H., Johansen H. K., Andersen L. P., Jarlov J. O., Boye K., Larsen A. R., Westh H. Comparing Whole-Genome Sequencing with Sanger Sequencing for spa Typing of Methicillin-Resistant Staphylococcus aureus. Journal of Clinical Microbiology. 2014;52(12):4305–4308. doi: 10.1128/JCM.01979-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laslett D, Bjorn C. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32(1):11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagesen Karin, Hallin Peter, Rødland Einar Andreas, Stærfeldt Hans-Henrik, Rognes Torbjørn, Ussery David W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camacho Christiam, Coulouris George, Avagyan Vahram, Ma Ning, Papadopoulos Jason, Bealer Kevin, Madden Thomas L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10(1):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC bioinformatics. 2010. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed]
- 21.Aziz RK, Bartels D, Best AA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatusova T, DiCuccio M, Badretdin A, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant J. R., Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Research. 2008;36(Web Server):W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura Koichiro, Stecher Glen, Peterson Daniel, Filipski Alan, Kumar Sudhir. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the current study are available from the corresponding author publicly available due (Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession NFZY00000000 and PRJNA385553. The version described in this paper is version NFZY01000000.